Deposition Date
2019-10-03
Release Date
2020-10-07
Last Version Date
2023-10-11
Entry Detail
PDB ID:
6UJI
Keywords:
Title:
Low resolution crystal structure (5.5 A) of the anthrax toxin protective antigen heptamer prepore D425A mutant
Biological Source:
Source Organism:
Bacillus anthracis (Taxon ID: 1392)
Host Organism:
Method Details:
Experimental Method:
Resolution:
5.50 Å
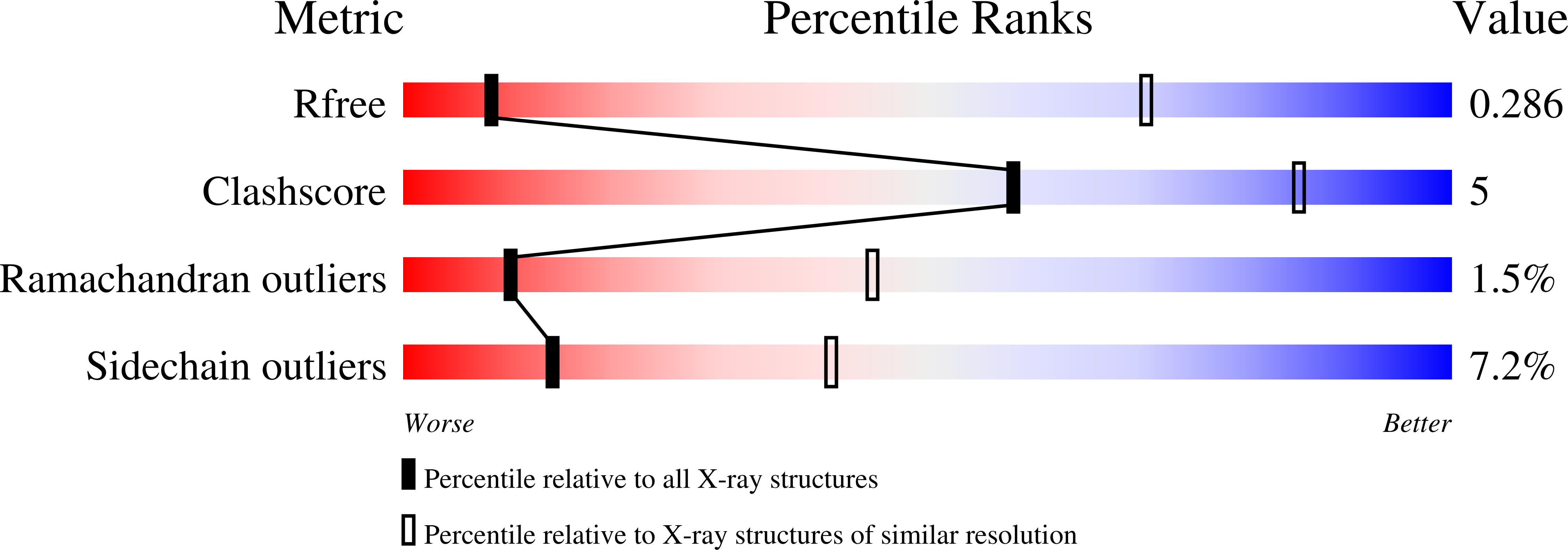
R-Value Free:
0.27
R-Value Work:
0.25
R-Value Observed:
0.25
Space Group:
P 1 21 1