Deposition Date
2019-09-24
Release Date
2020-03-18
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6UFP
Keywords:
Title:
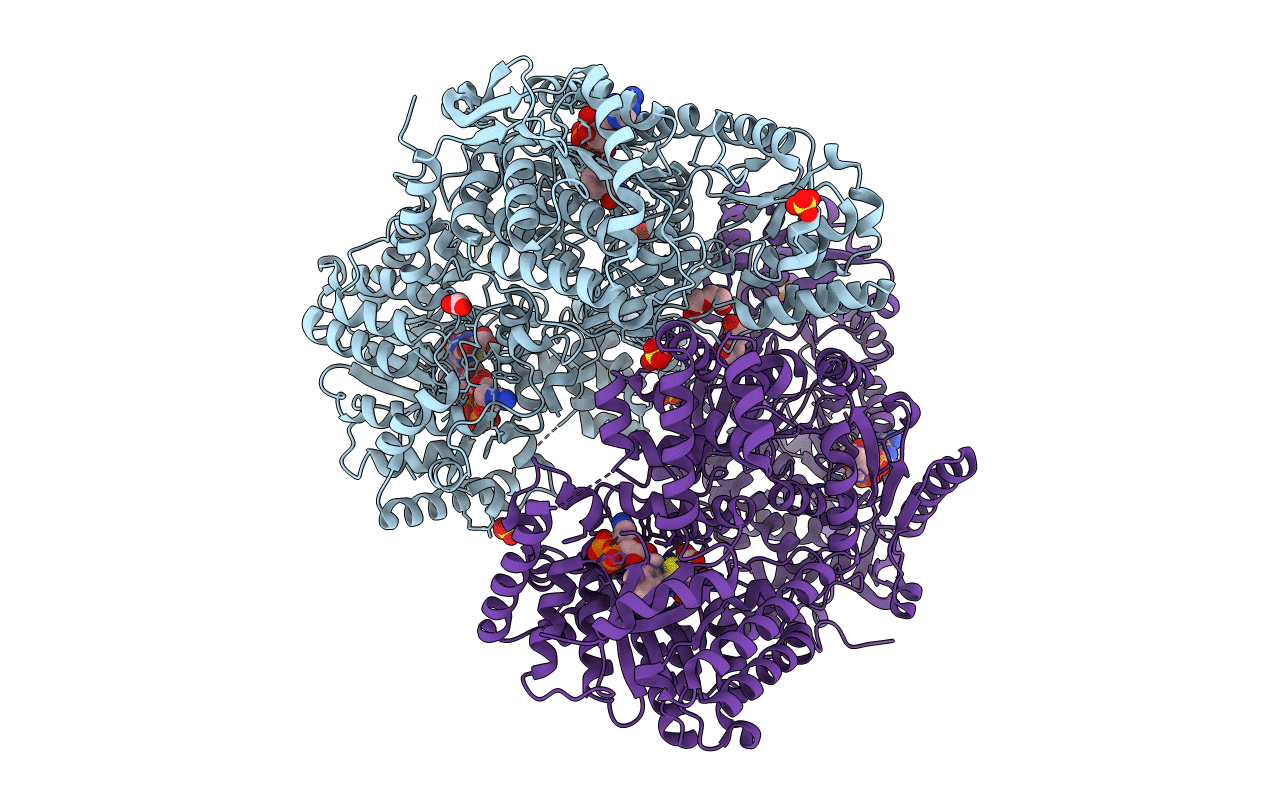
Structure of proline utilization A with the FAD covalently modified by L-thiazolidine-2-carboxylate and three cysteines (Cys46, Cys470, Cys638) modified to S,S-(2-HYDROXYETHYL)THIOCYSTEINE
Biological Source:
Source Organism(s):
Sinorhizobium meliloti (strain SM11) (Taxon ID: 707241)
Expression System(s):
Method Details:
Experimental Method:
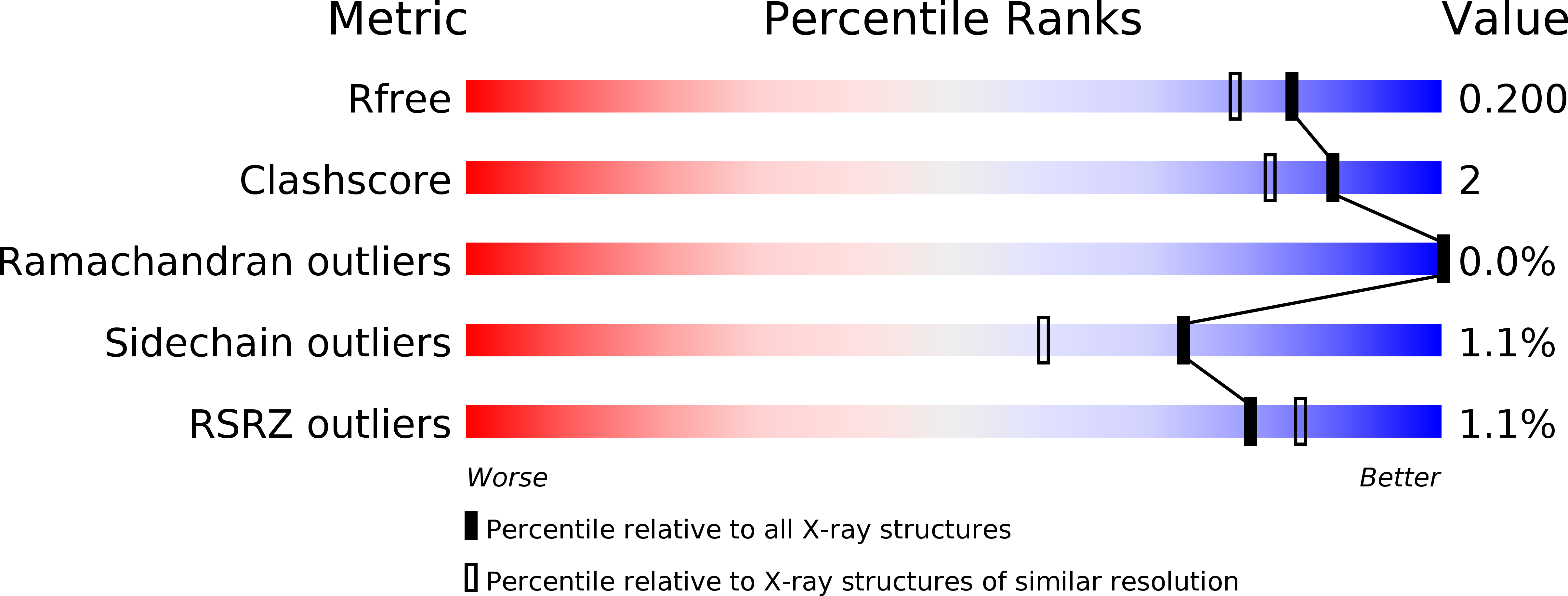
Resolution:
1.74 Å
R-Value Free:
0.19
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 1 21 1