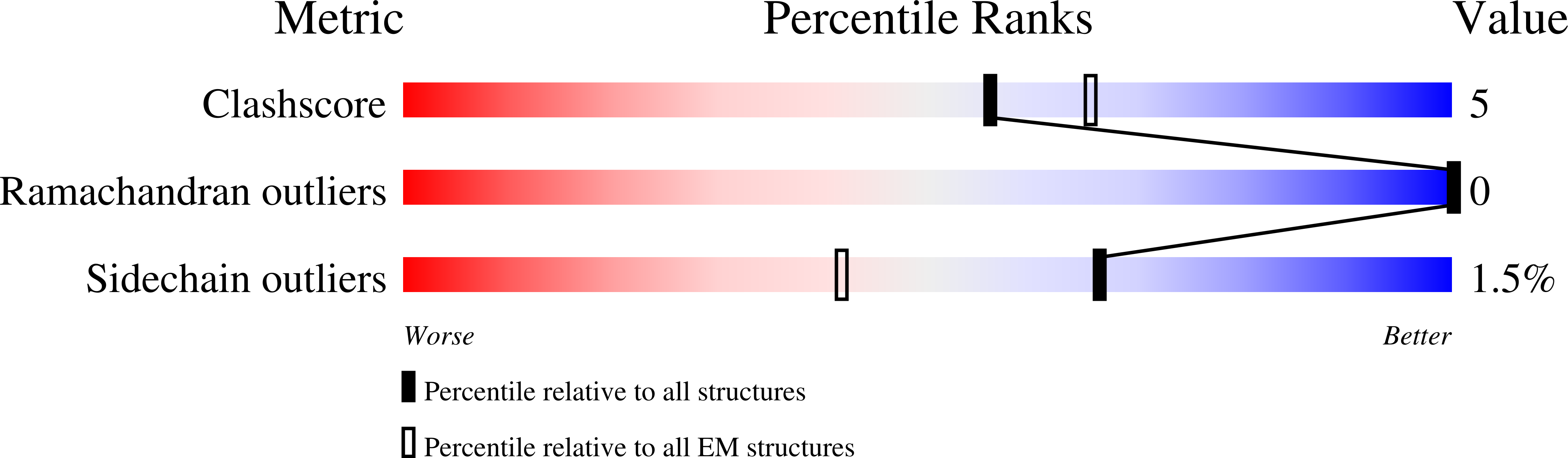
Deposition Date
2019-08-27
Release Date
2020-04-15
Last Version Date
2024-03-20
Entry Detail
PDB ID:
6U5K
Keywords:
Title:
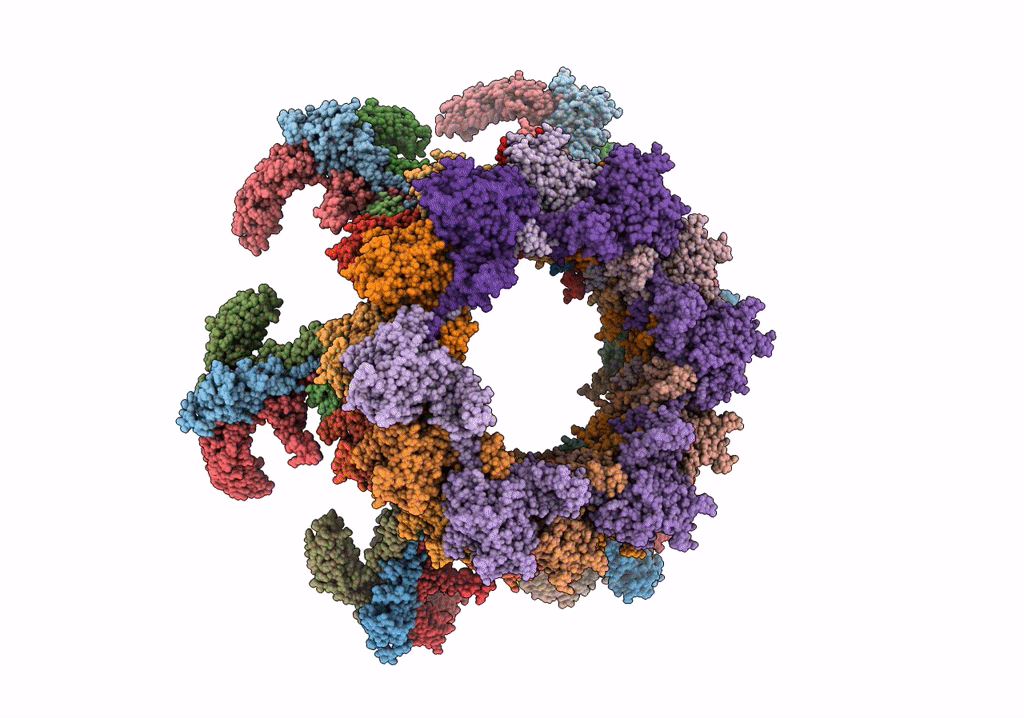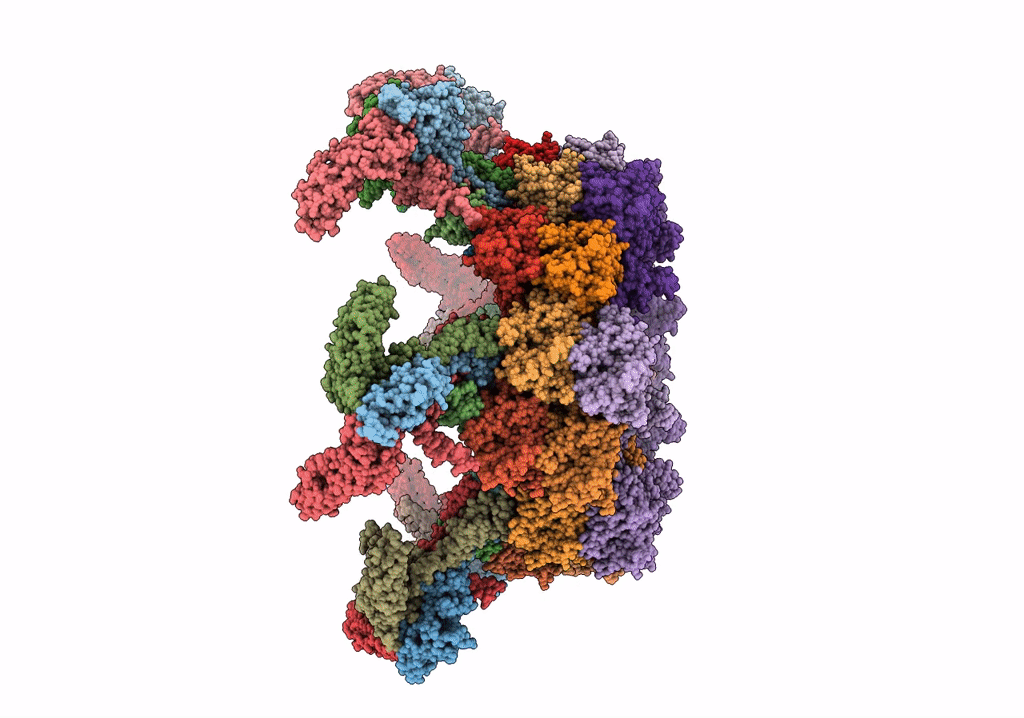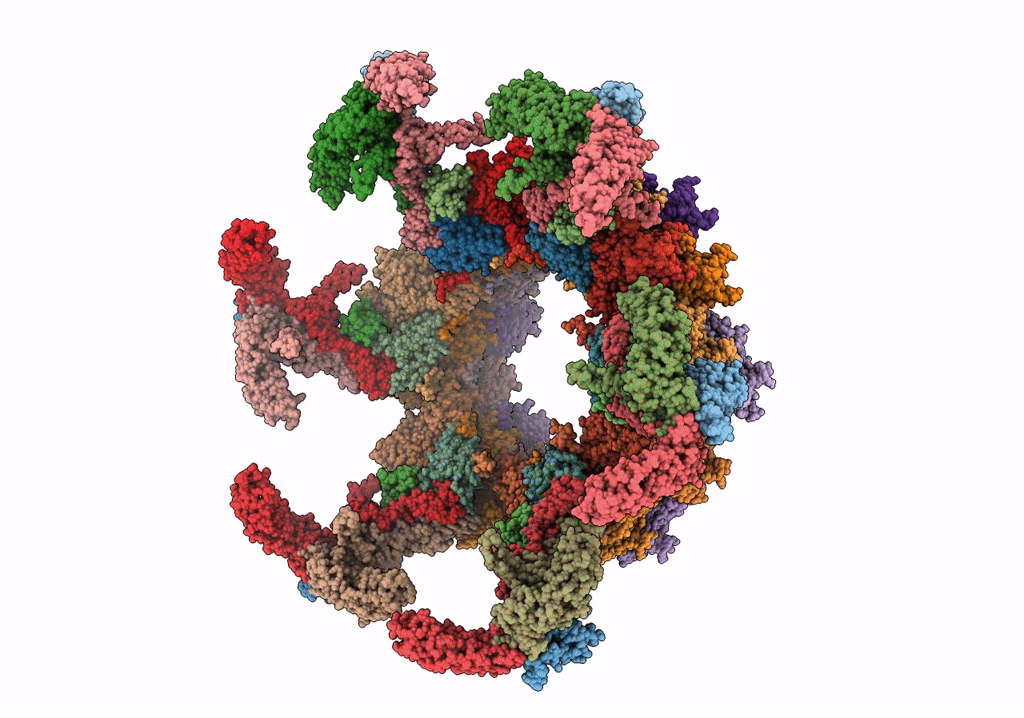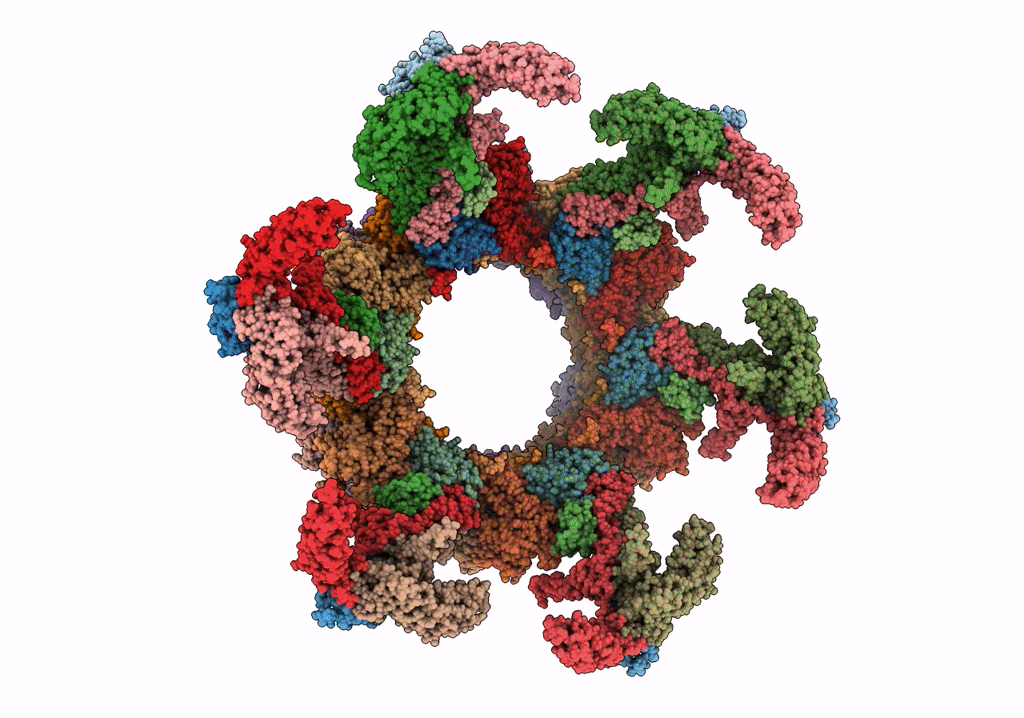
CryoEM Structure of Pyocin R2 - postcontracted - baseplate
Biological Source:
Source Organism(s):
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE