Deposition Date
2020-01-09
Release Date
2020-04-22
Last Version Date
2024-01-24
Entry Detail
PDB ID:
6TV9
Keywords:
Title:
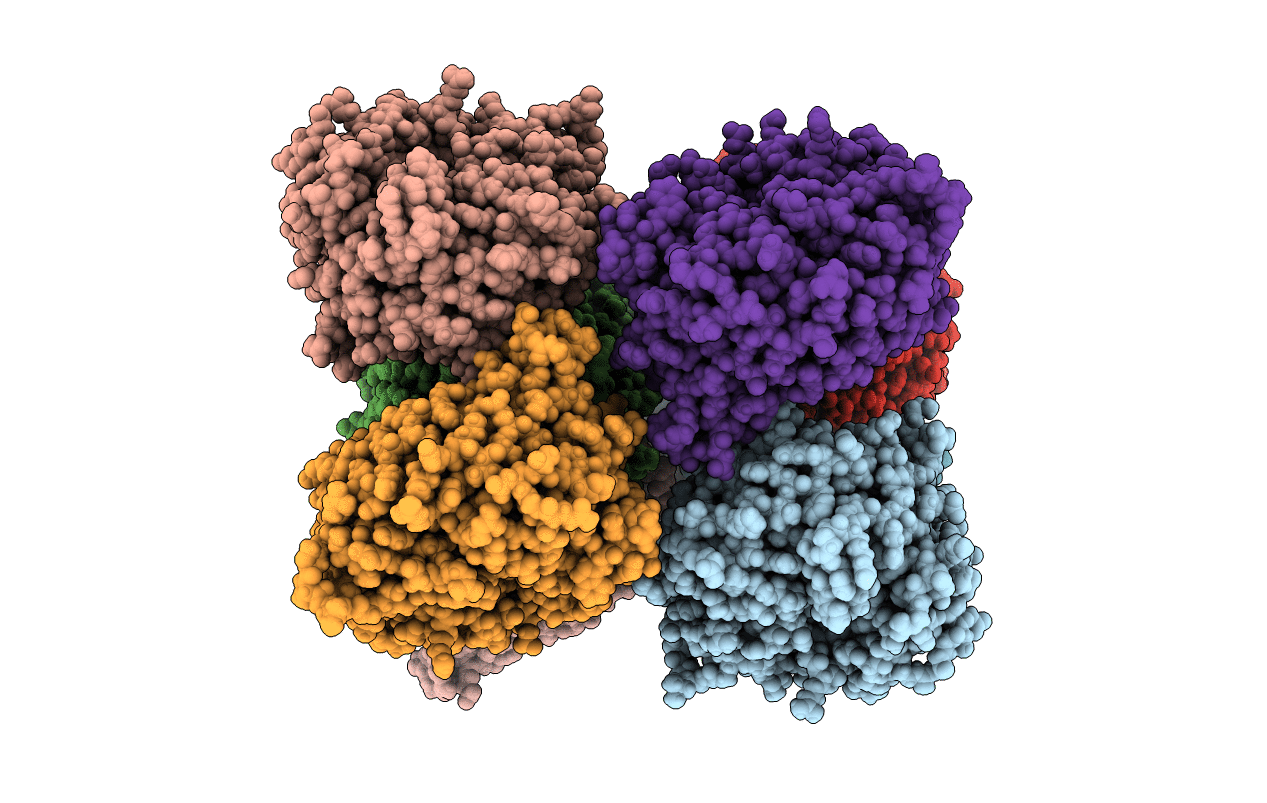
Heme d1 biosynthesis associated Protein NirF in complex with dihydro-heme d1
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
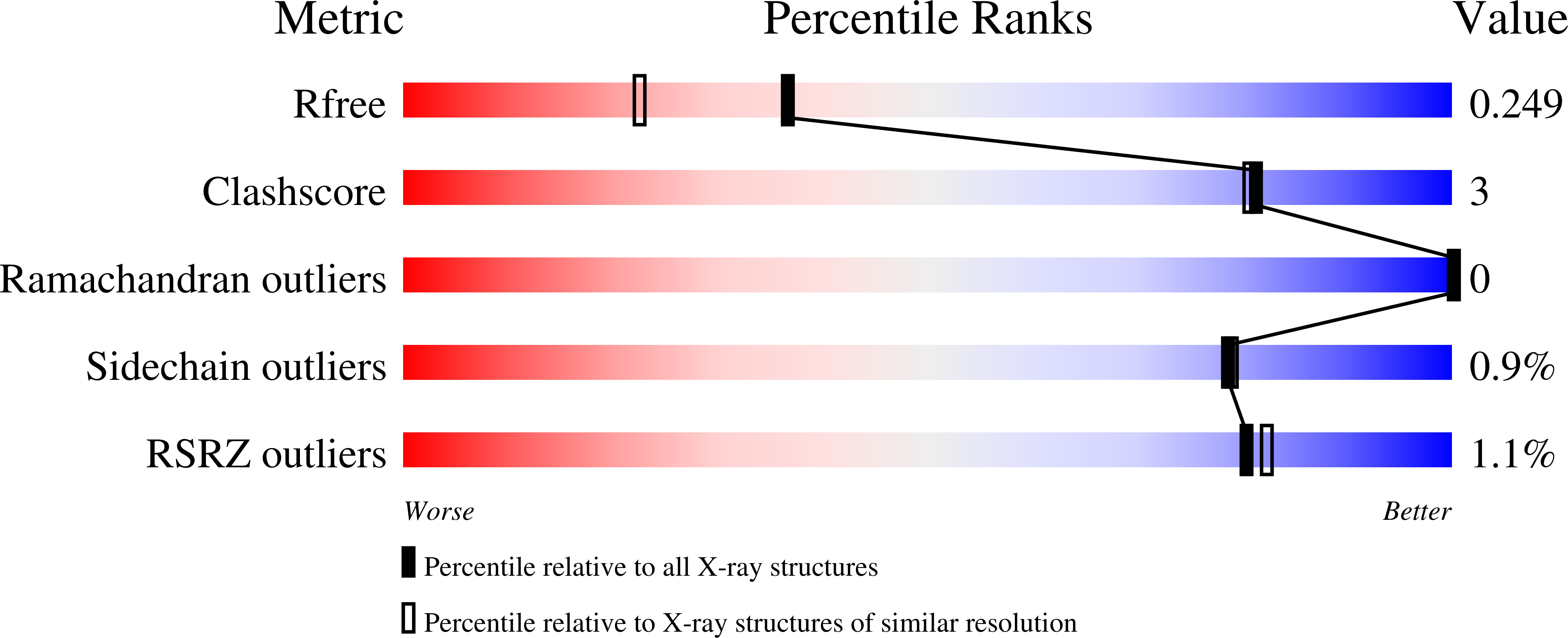
Resolution:
1.89 Å
R-Value Free:
0.25
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 1 21 1