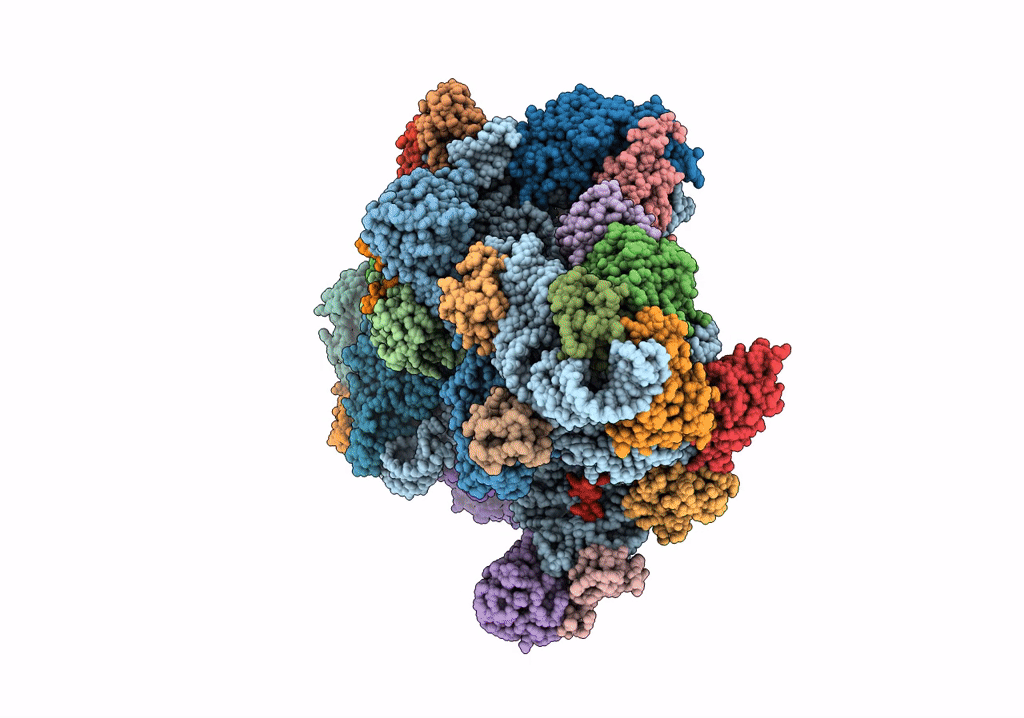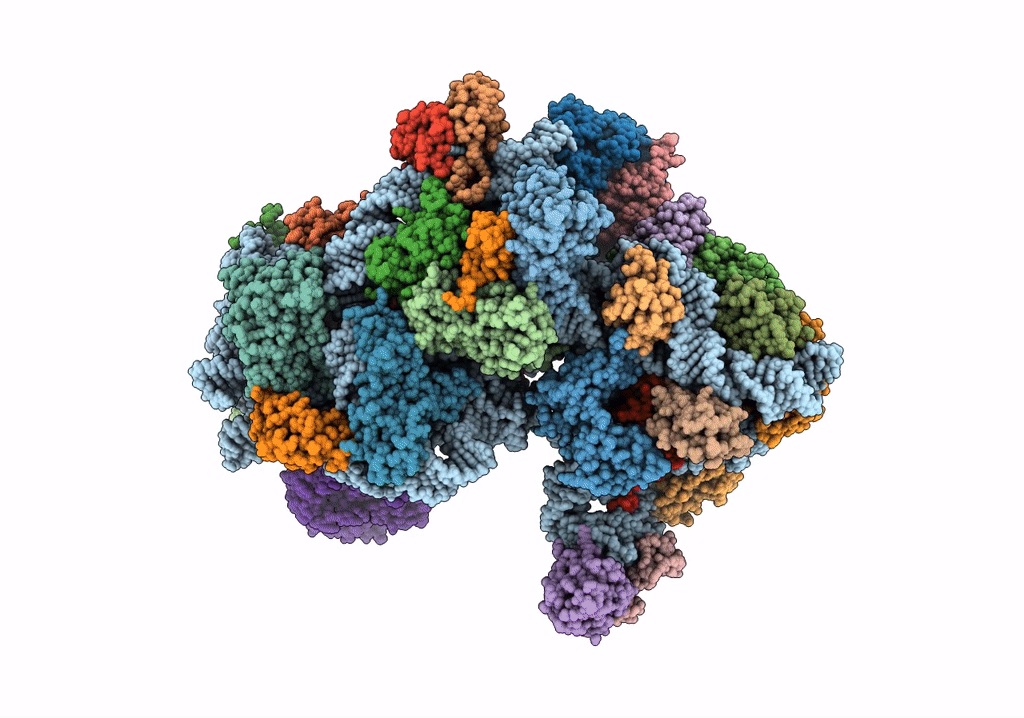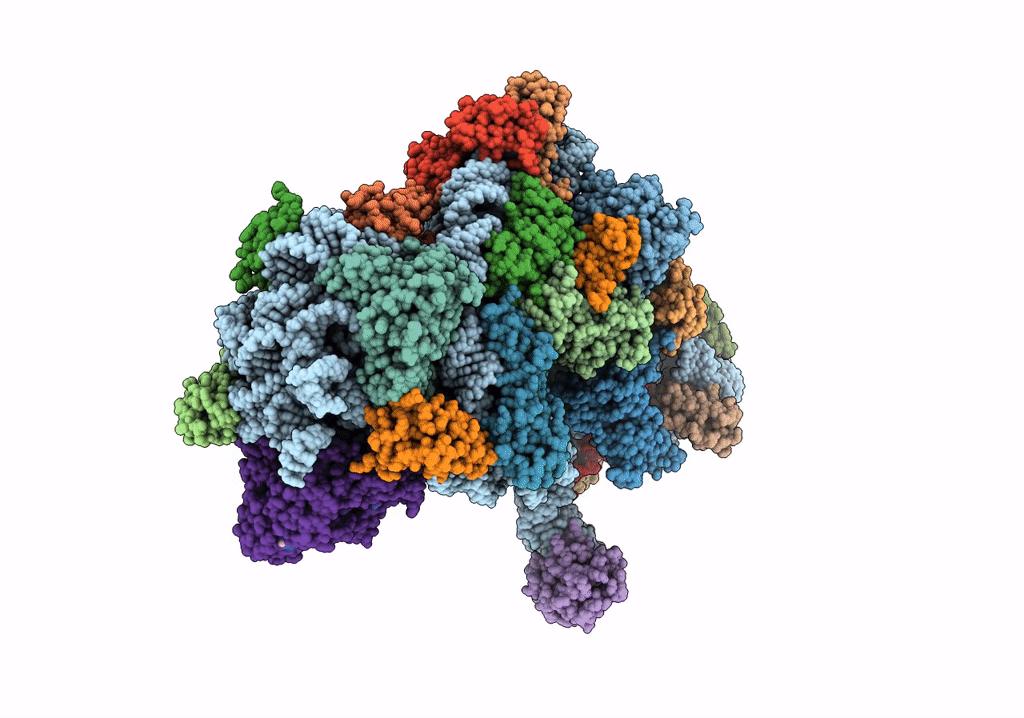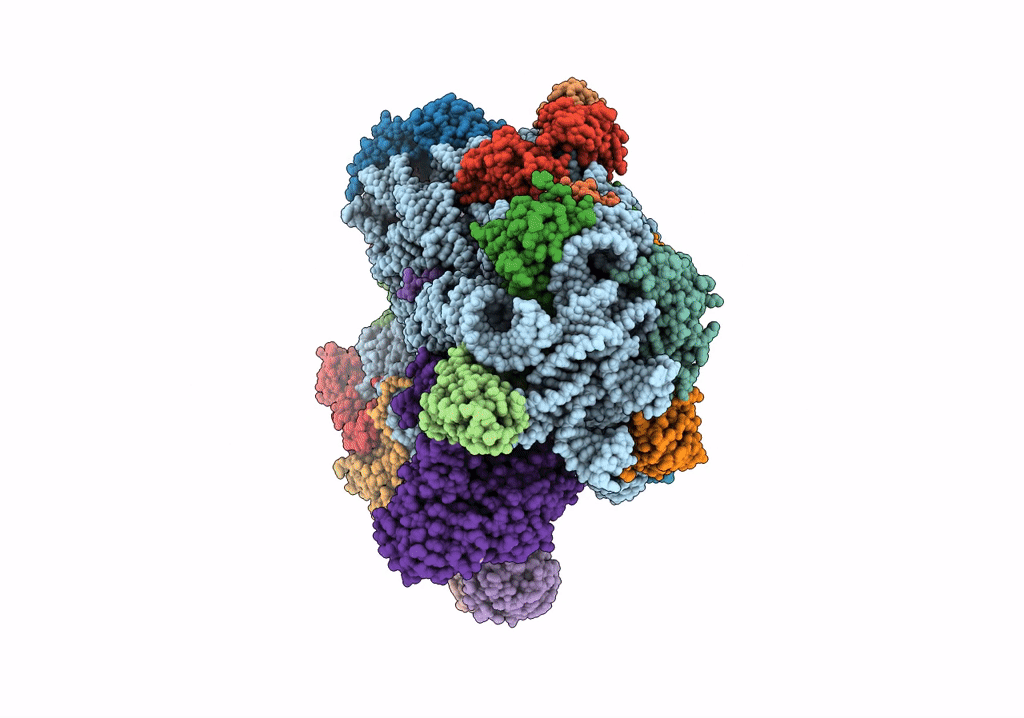
Deposition Date
2019-12-04
Release Date
2020-02-12
Last Version Date
2024-05-22
Entry Detail
PDB ID:
6TMF
Keywords:
Title:
Structure of an archaeal ABCE1-bound ribosomal post-splitting complex
Biological Source:
Source Organism(s):
Saccharolobus solfataricus (Taxon ID: 2287)
Thermococcus celer Vu 13 = JCM 8558 (Taxon ID: 1293037)
Thermococcus celer Vu 13 = JCM 8558 (Taxon ID: 1293037)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


