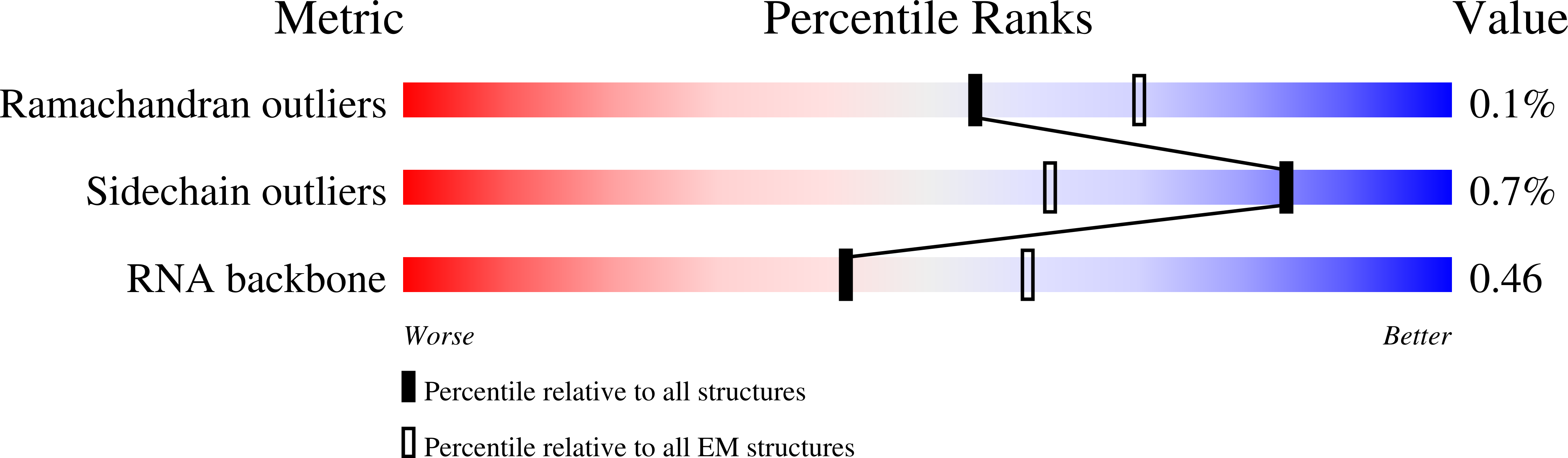
Deposition Date
2019-10-15
Release Date
2019-11-27
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6T59
Keywords:
Title:
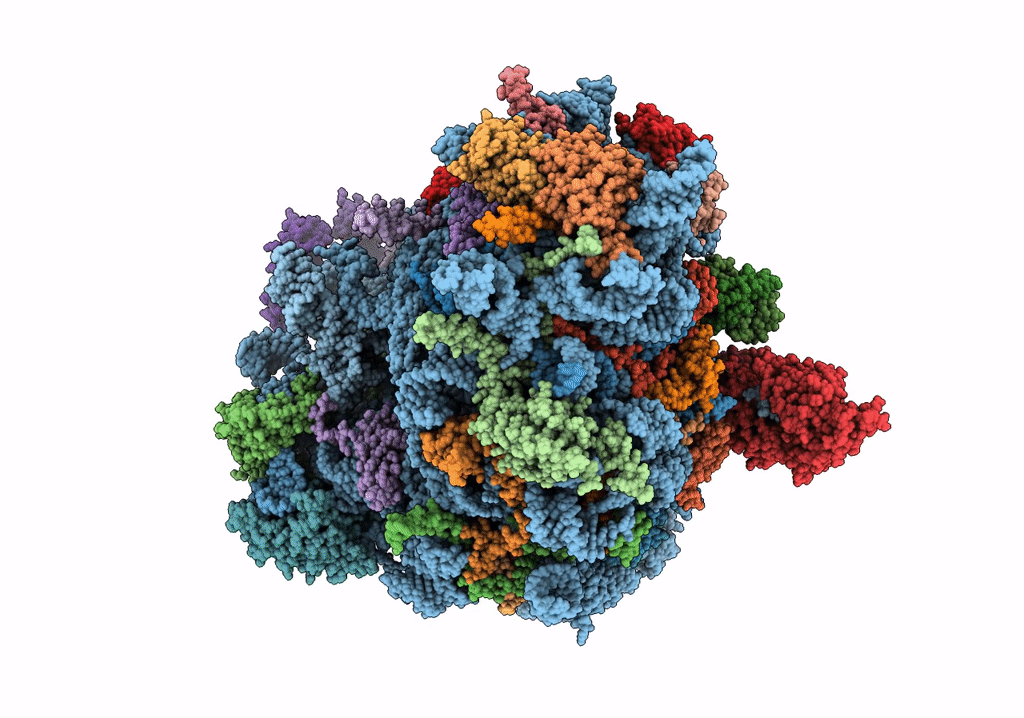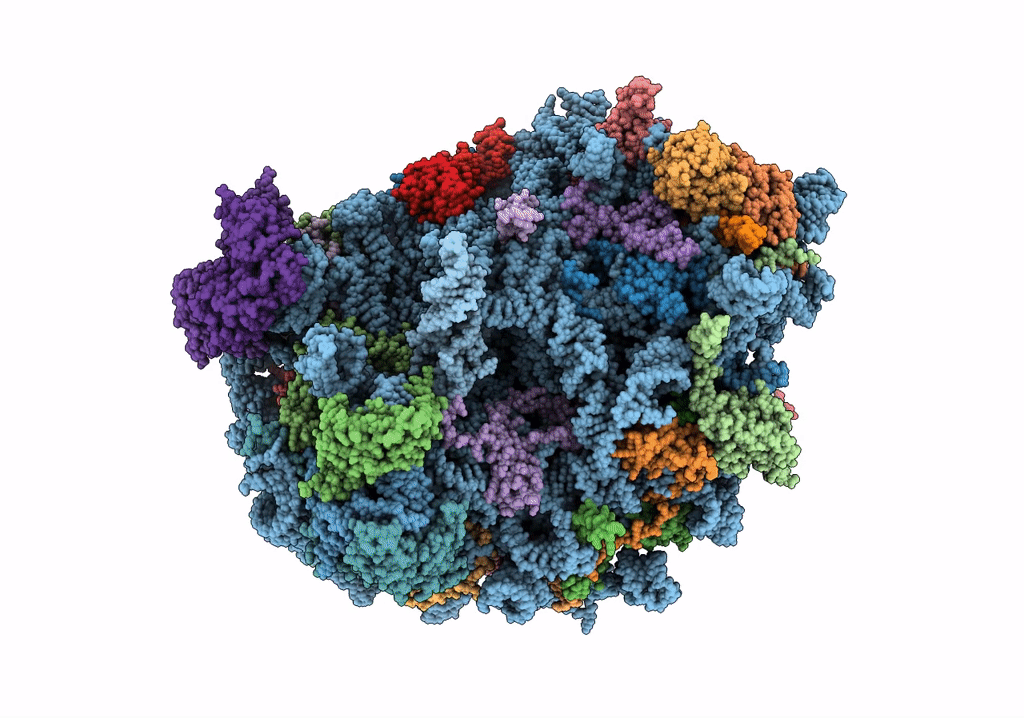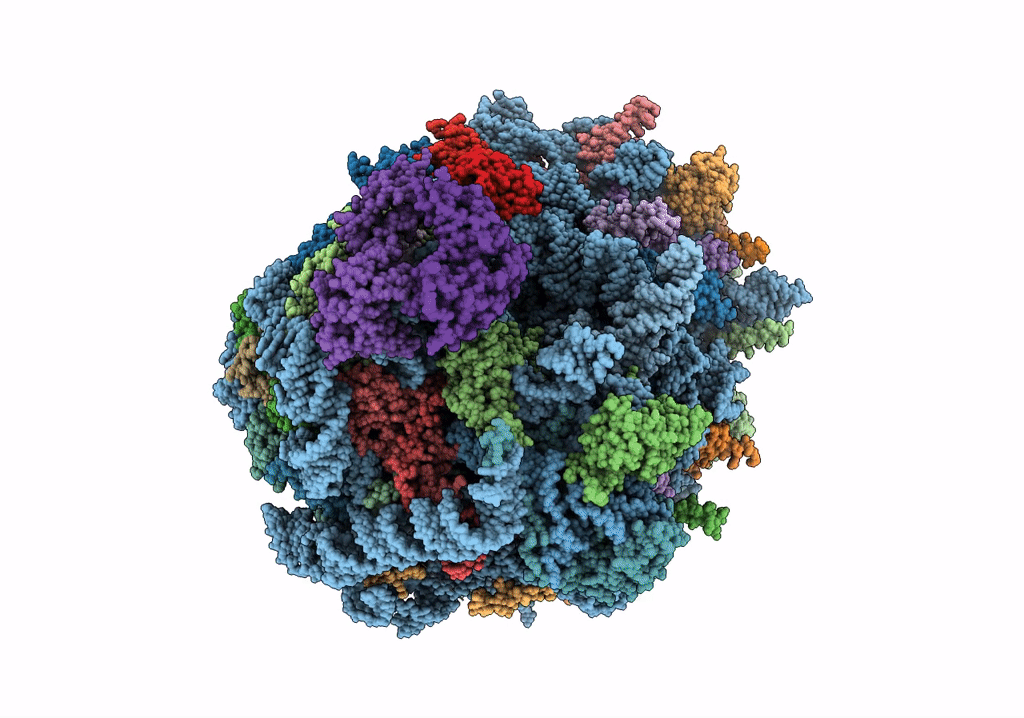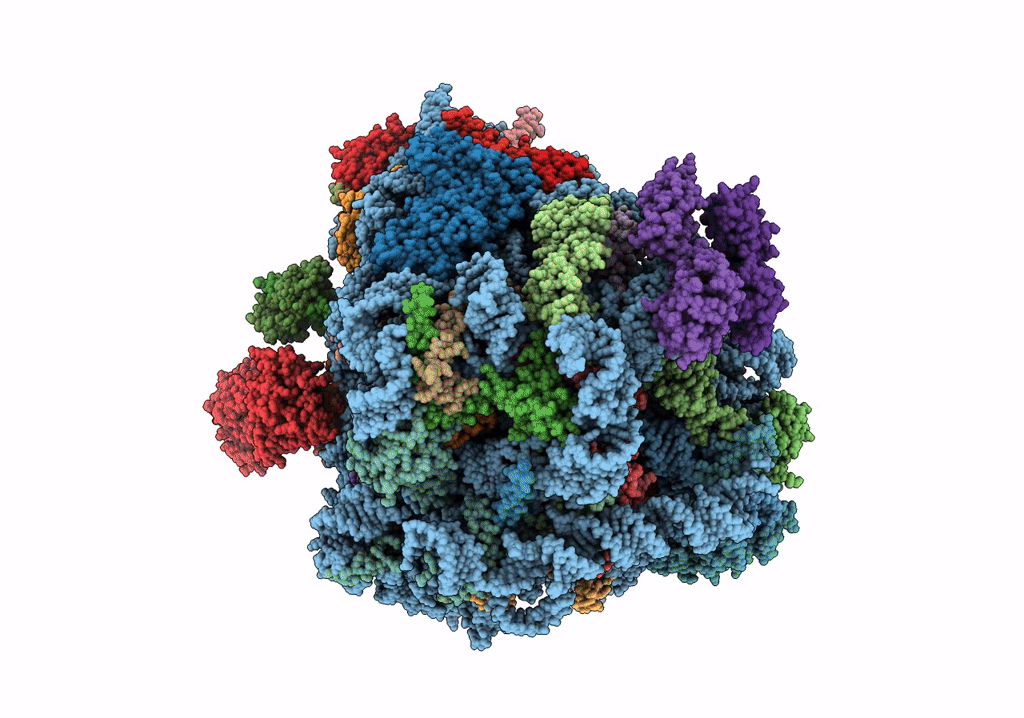
Structure of rabbit 80S ribosome translating beta-tubulin in complex with tetratricopeptide protein 5 and nascent chain-associated complex
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Oryctolagus cuniculus (Taxon ID: 9986)
Oryctolagus cuniculus (Taxon ID: 9986)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.11 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE