Deposition Date
2019-09-26
Release Date
2020-02-26
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6SXR
Keywords:
Title:
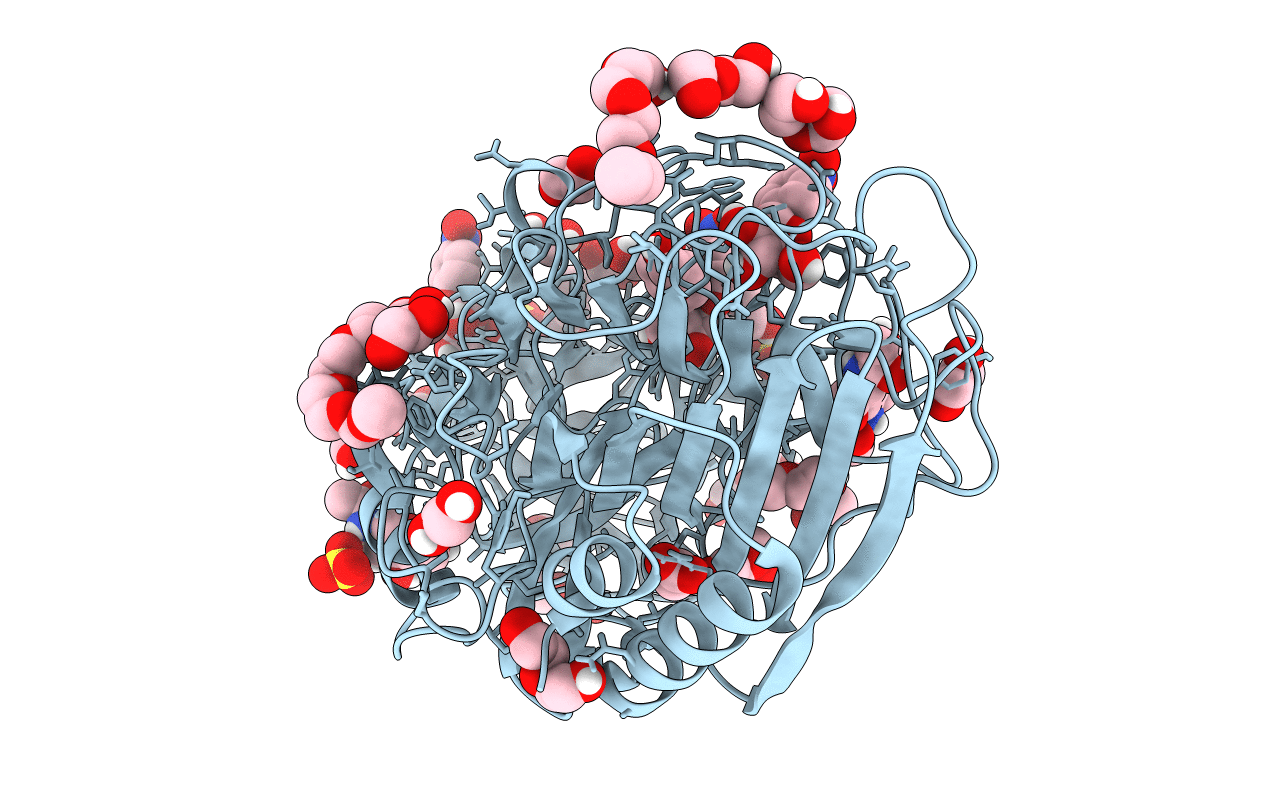
E221Q mutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside
Biological Source:
Source Organism(s):
Aspergillus kawachii IFO 4308 (Taxon ID: 1033177)
Expression System(s):
Method Details:
Experimental Method:
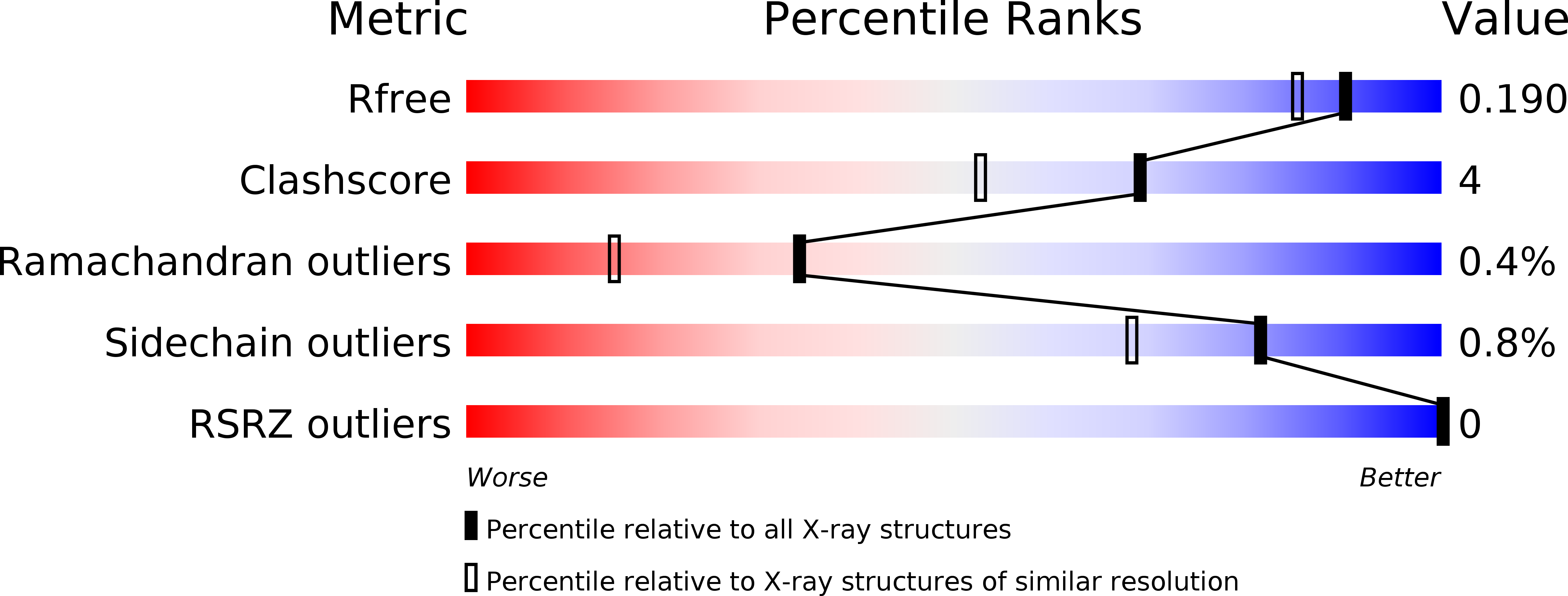
Resolution:
1.64 Å
R-Value Free:
0.18
R-Value Work:
0.15
Space Group:
H 3 2