Deposition Date
2019-09-12
Release Date
2019-12-11
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6SU3
Keywords:
Title:
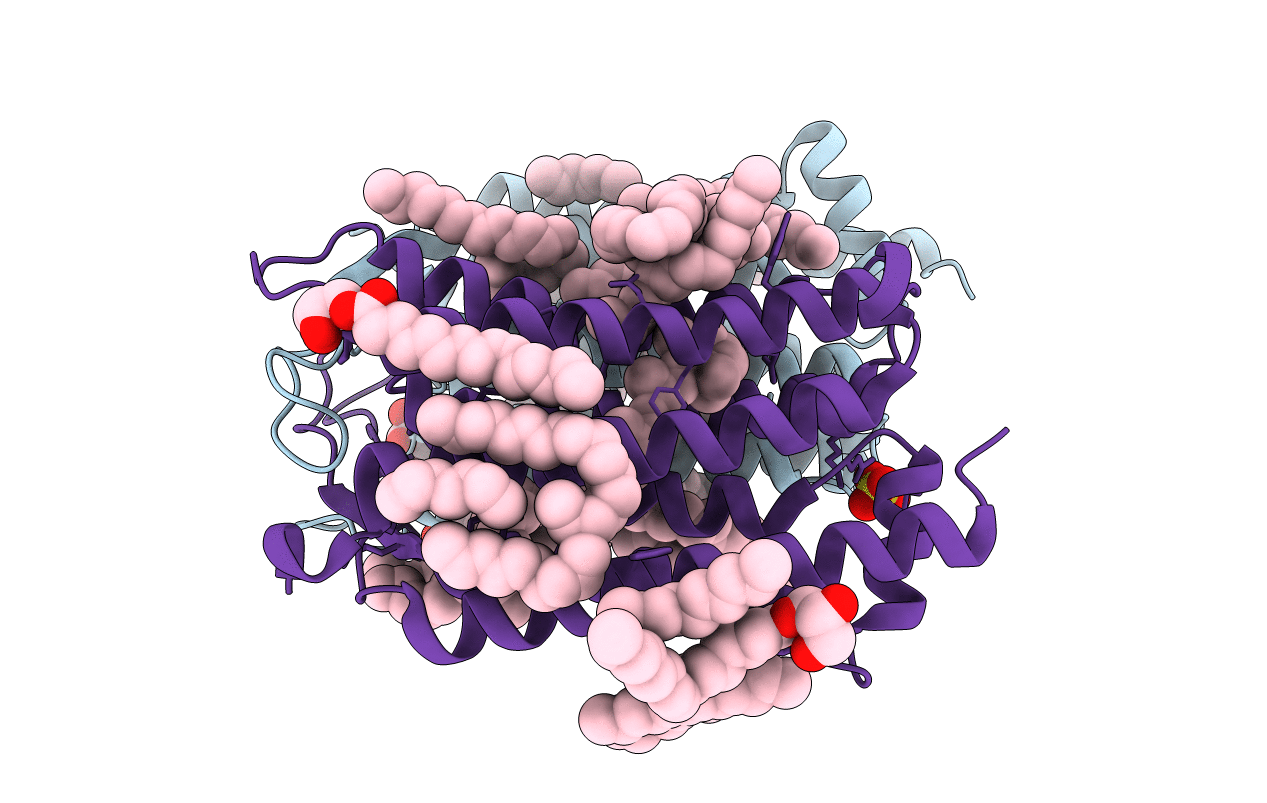
Crystal structure of the 48C12 heliorhodopsin in the violet form at pH 8.8
Biological Source:
Source Organism:
Actinobacteria bacterium (Taxon ID: 1883427)
Host Organism:
Method Details:
Experimental Method:
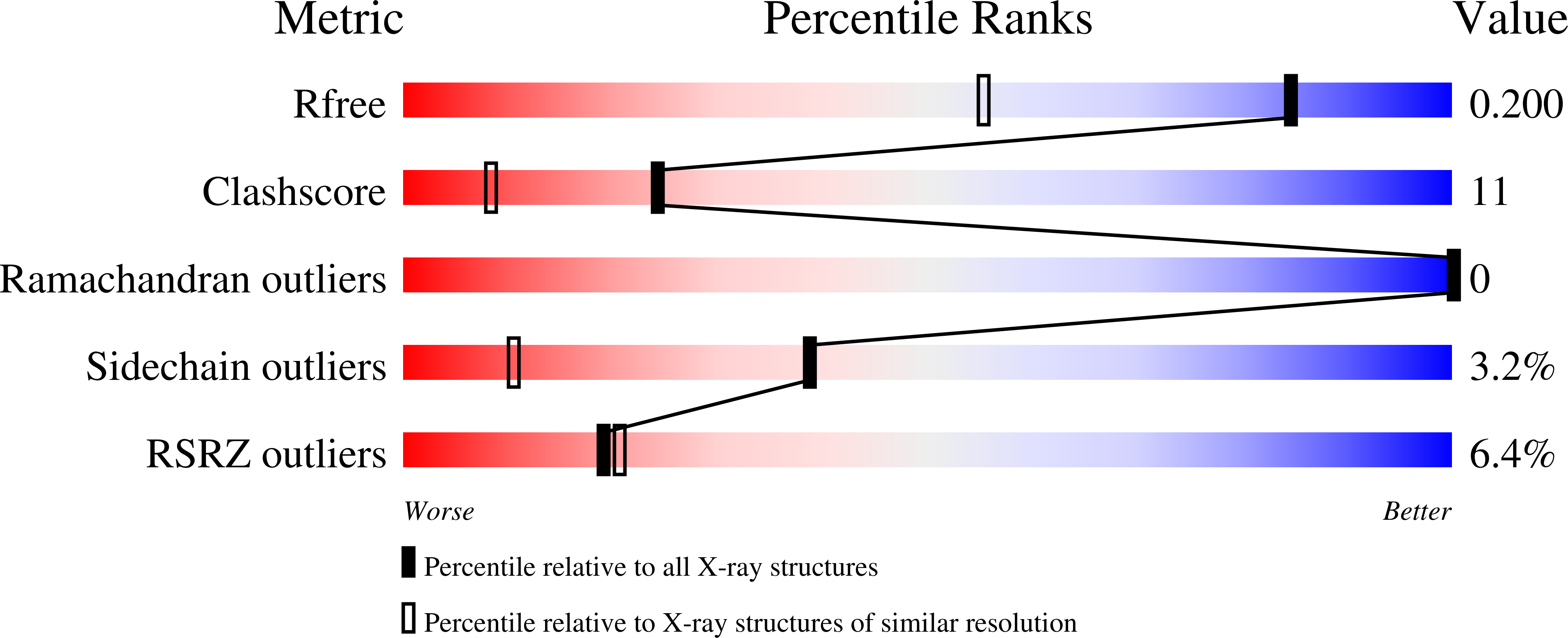
Resolution:
1.50 Å
R-Value Free:
0.19
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
P 1 21 1