Deposition Date
2019-07-30
Release Date
2019-09-25
Last Version Date
2024-05-15
Entry Detail
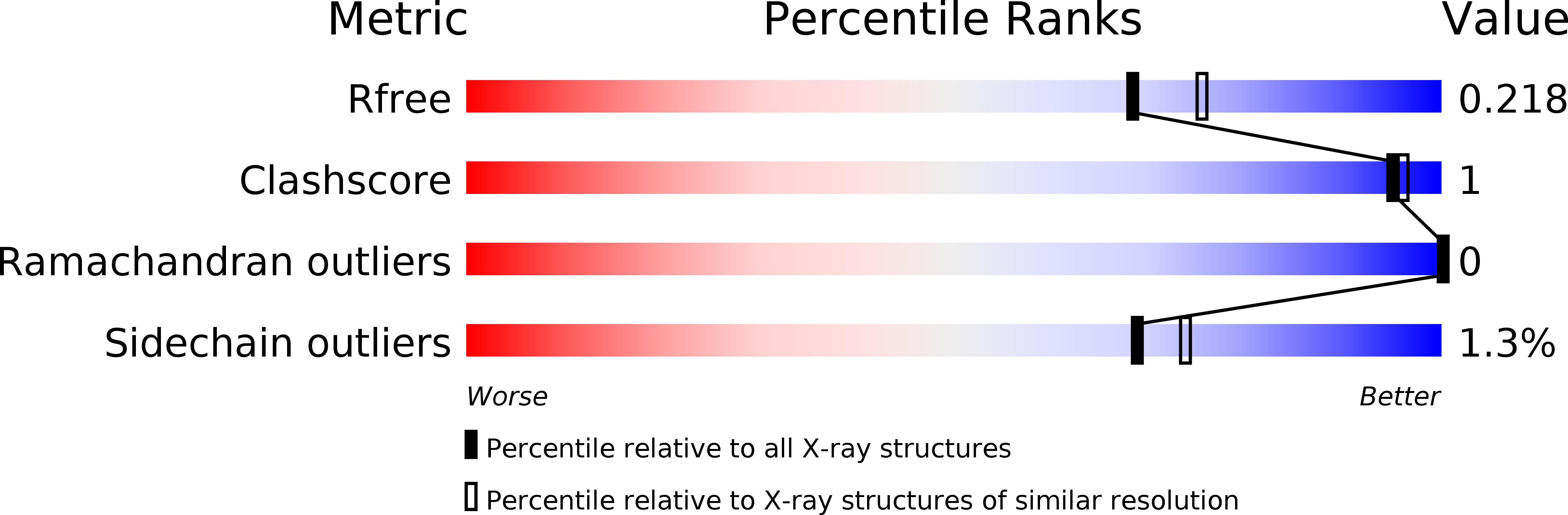
PDB ID:
6SEV
Keywords:
Title:
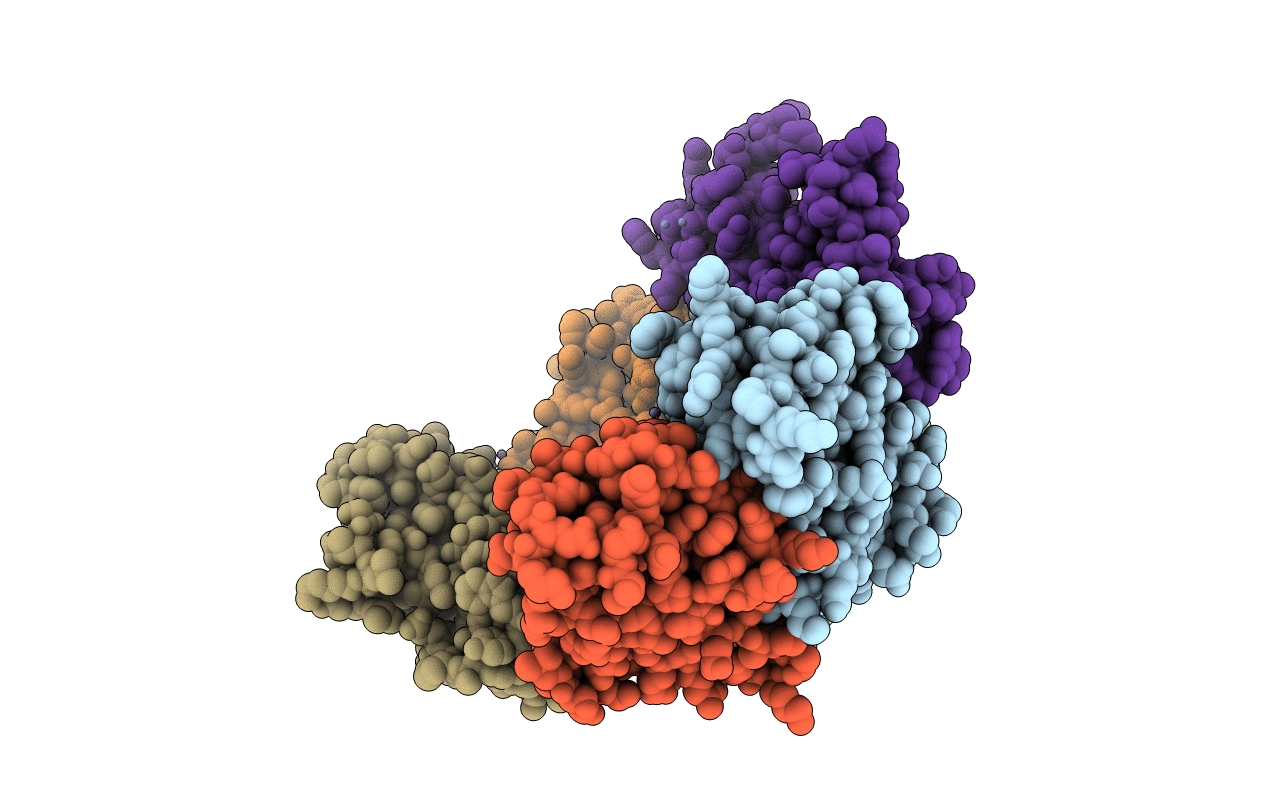
Structure of Dps from Listeria innocua soaked with 10 mM zinc for 120 minutes
Biological Source:
Source Organism(s):
Listeria innocua (Taxon ID: 1642)
Expression System(s):
Method Details: