Deposition Date
2019-06-14
Release Date
2019-06-26
Last Version Date
2024-01-24
Entry Detail
PDB ID:
6S07
Keywords:
Title:
Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen.
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.04 Å
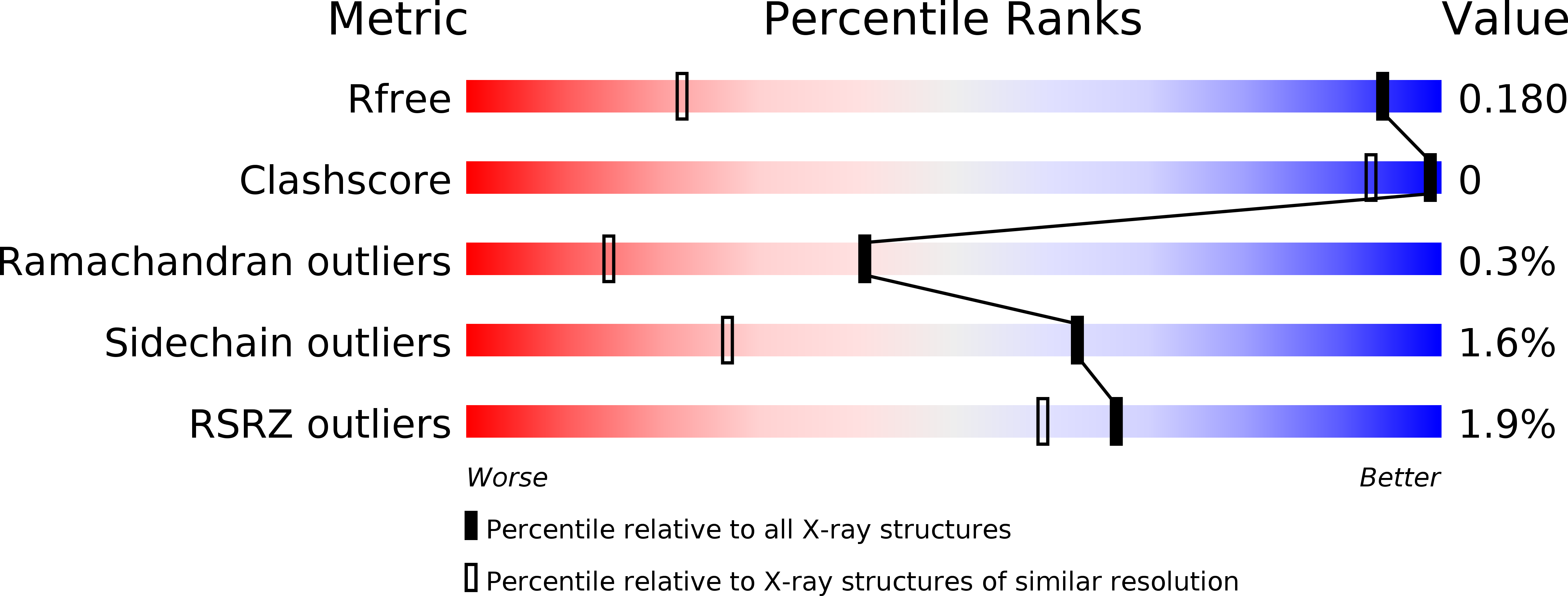
R-Value Free:
0.18
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 21 21 21