Deposition Date
2019-05-21
Release Date
2019-09-18
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6RSL
Keywords:
Title:
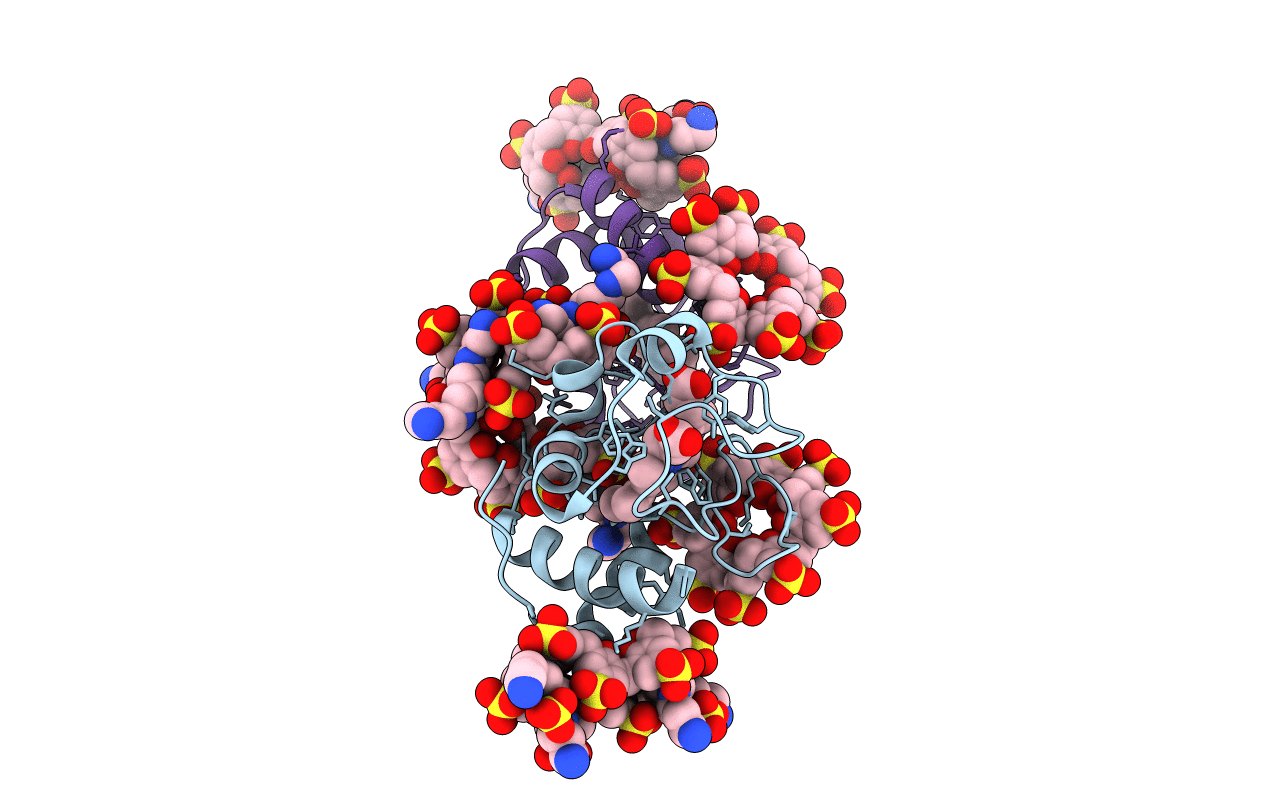
Cytochrome c co-crystallized with 10 eq. sulfonato-calix[8]arene and 25 eq. spermine (dry-coating method) - structure III
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
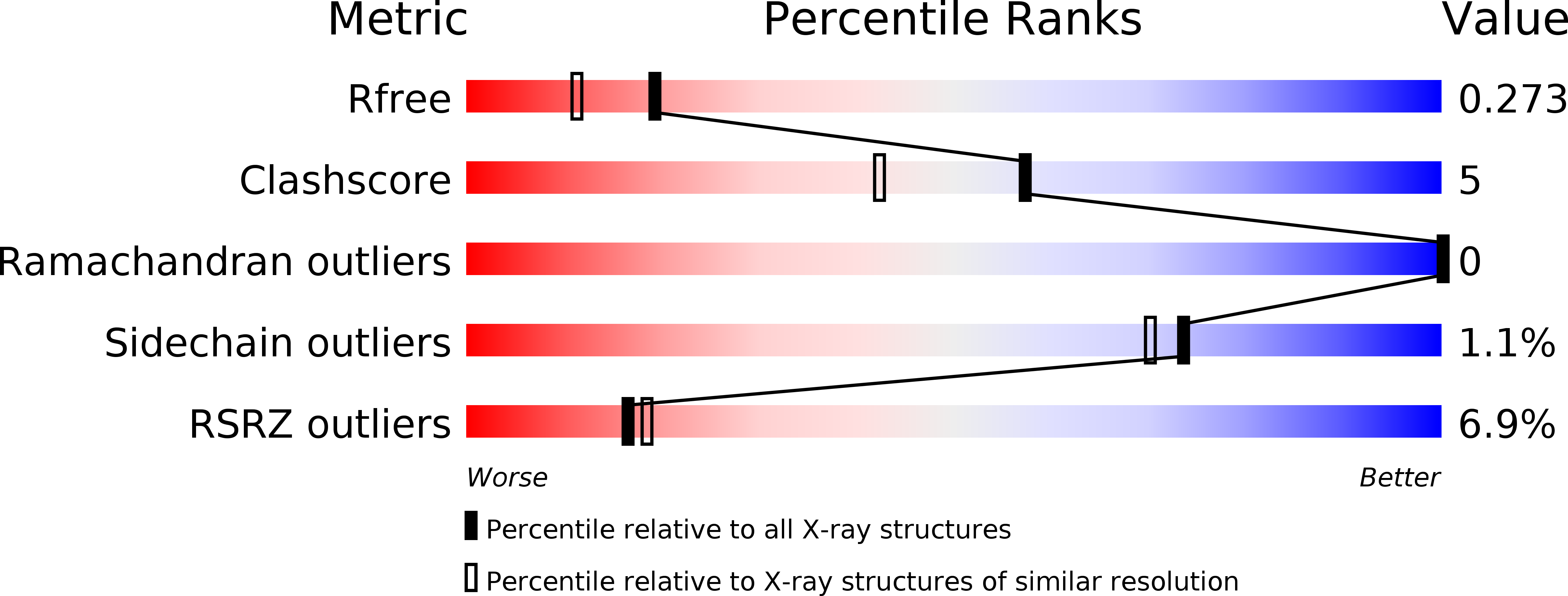
Resolution:
1.99 Å
R-Value Free:
0.26
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 43 21 2