Deposition Date
2019-04-23
Release Date
2019-05-01
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6RI0
Keywords:
Title:
Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose.
Biological Source:
Source Organism(s):
Steccherinum murashkinskyi (Taxon ID: 627145)
Method Details:
Experimental Method:
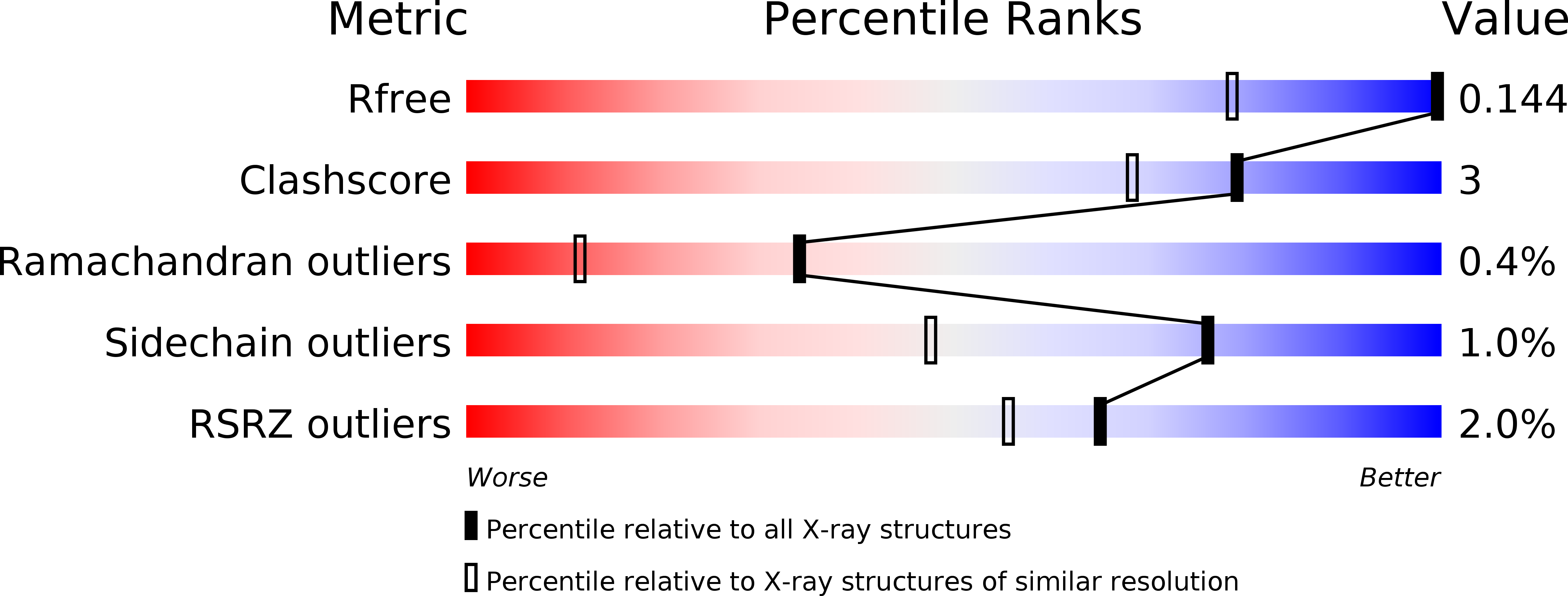
Resolution:
1.00 Å
R-Value Free:
0.13
R-Value Work:
0.11
R-Value Observed:
0.12
Space Group:
P 21 21 21