Deposition Date
2018-12-31
Release Date
2019-04-24
Last Version Date
2024-05-15
Entry Detail
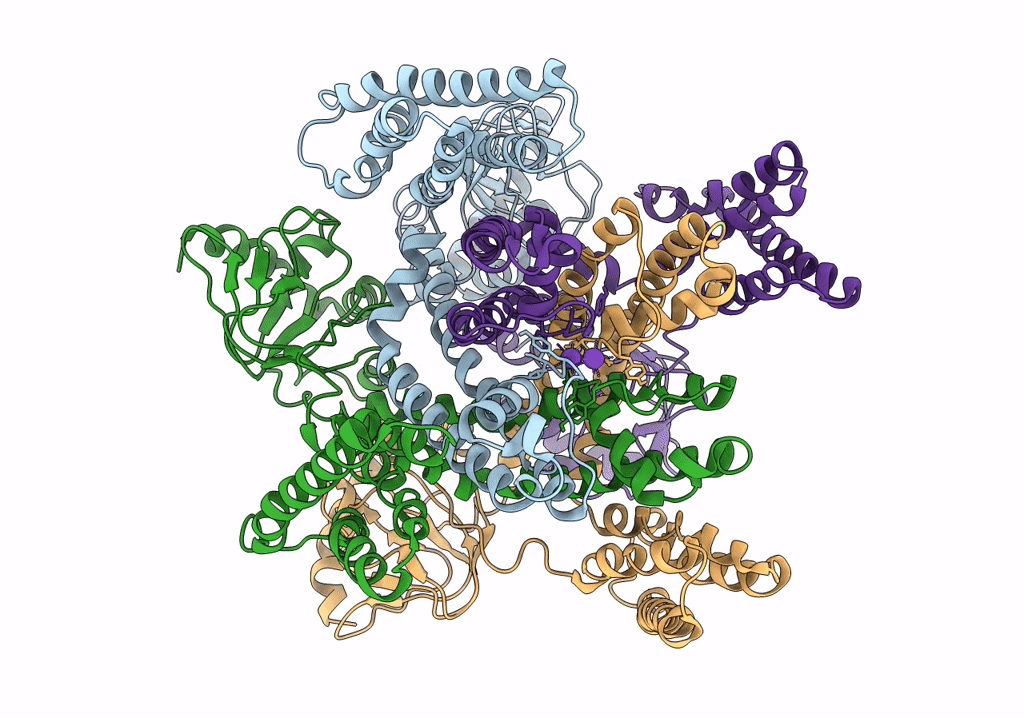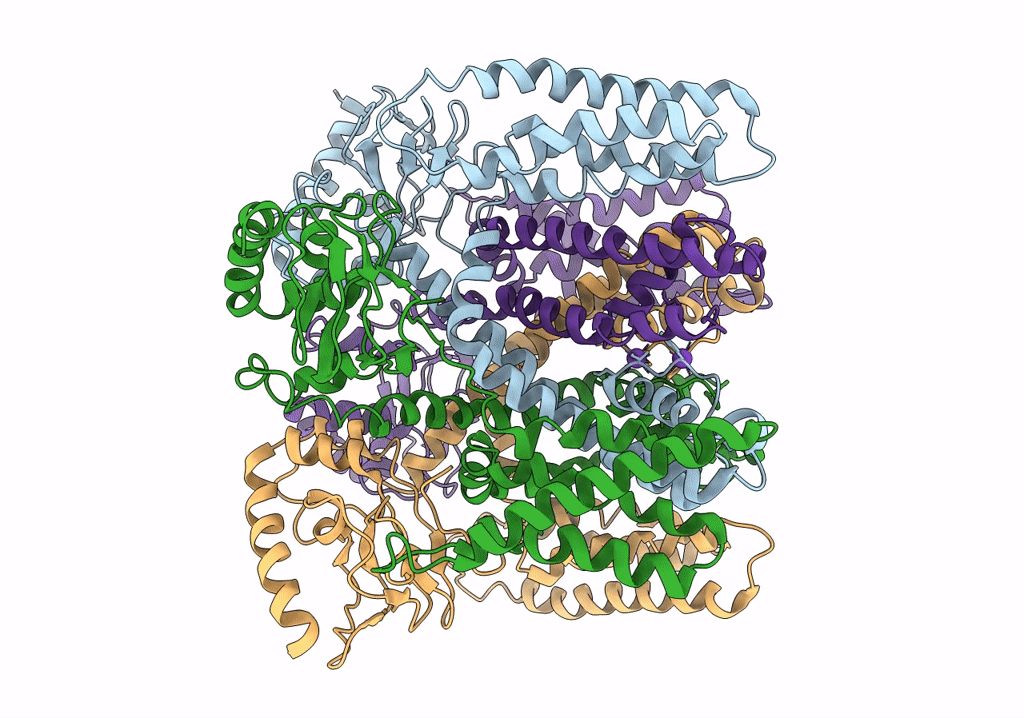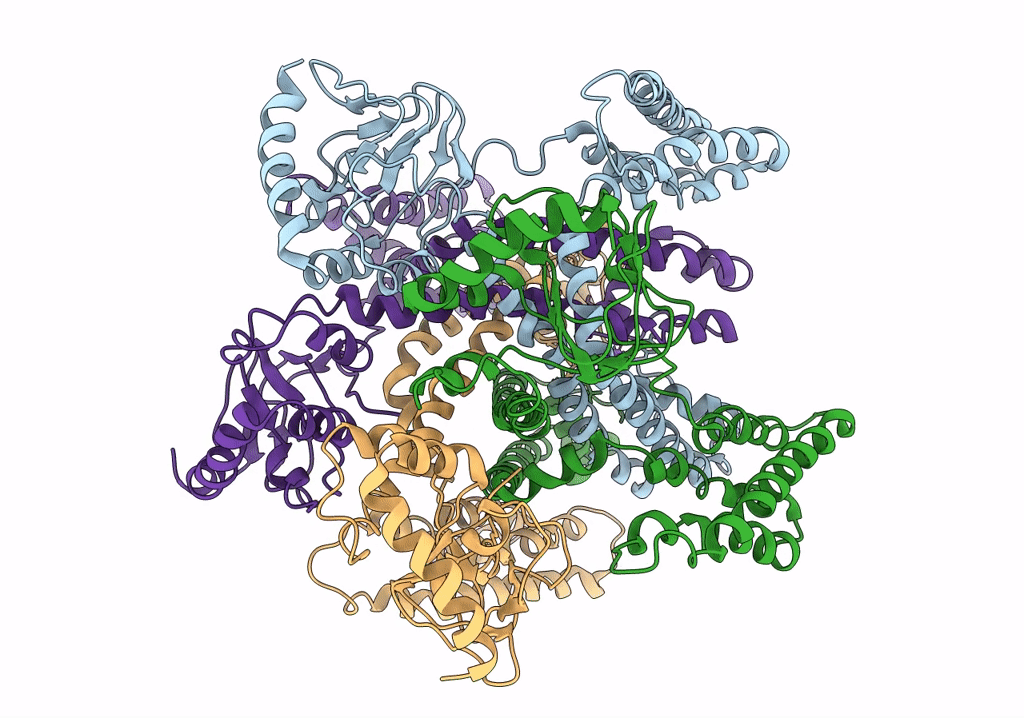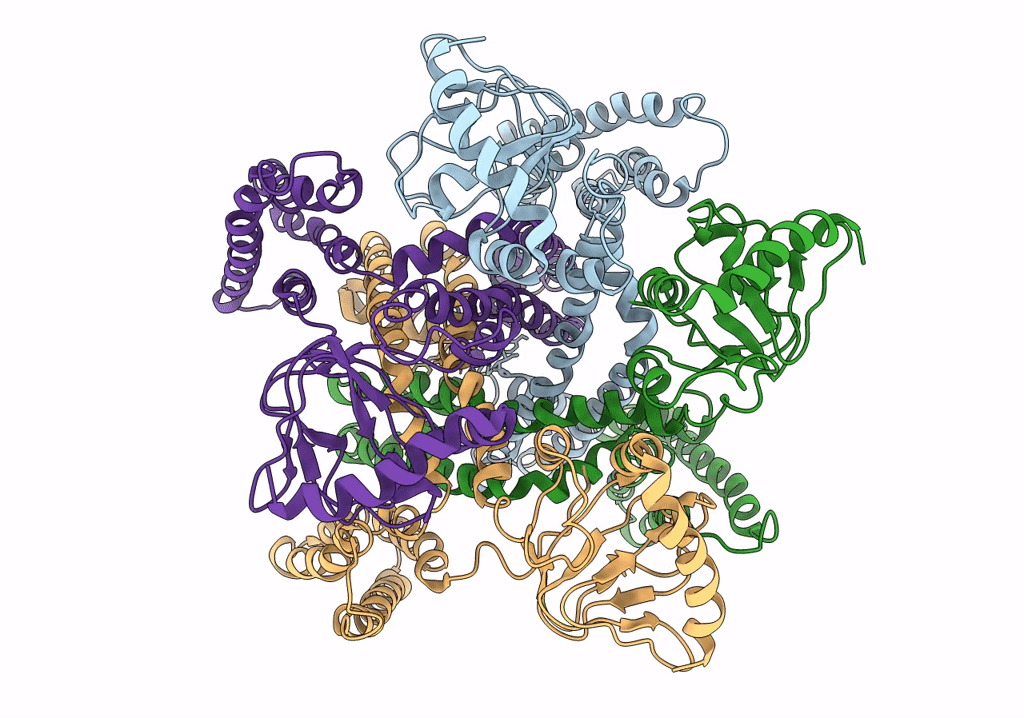
PDB ID:
6QD2
Keywords:
Title:
MloK1 model from single particle analysis of 2D crystals, class 6 (intermediate compact conformation)
Biological Source:
Source Organism(s):
Mesorhizobium loti MAFF303099 (Taxon ID: 266835)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.80 Å
Aggregation State:
2D ARRAY
Reconstruction Method:
SINGLE PARTICLE


