Deposition Date
2019-08-01
Release Date
2019-12-18
Last Version Date
2024-11-20
Entry Detail
PDB ID:
6Q0E
Keywords:
Title:
Inferred precursor (UCA) of the human antibody lineage 652 in complex with influenza hemagglutinin head domain of A/Beijing/262/95(H1N1)
Biological Source:
Source Organism:
Influenza A virus (A/Beijing/262/1995(H1N1)) (Taxon ID: 518922)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
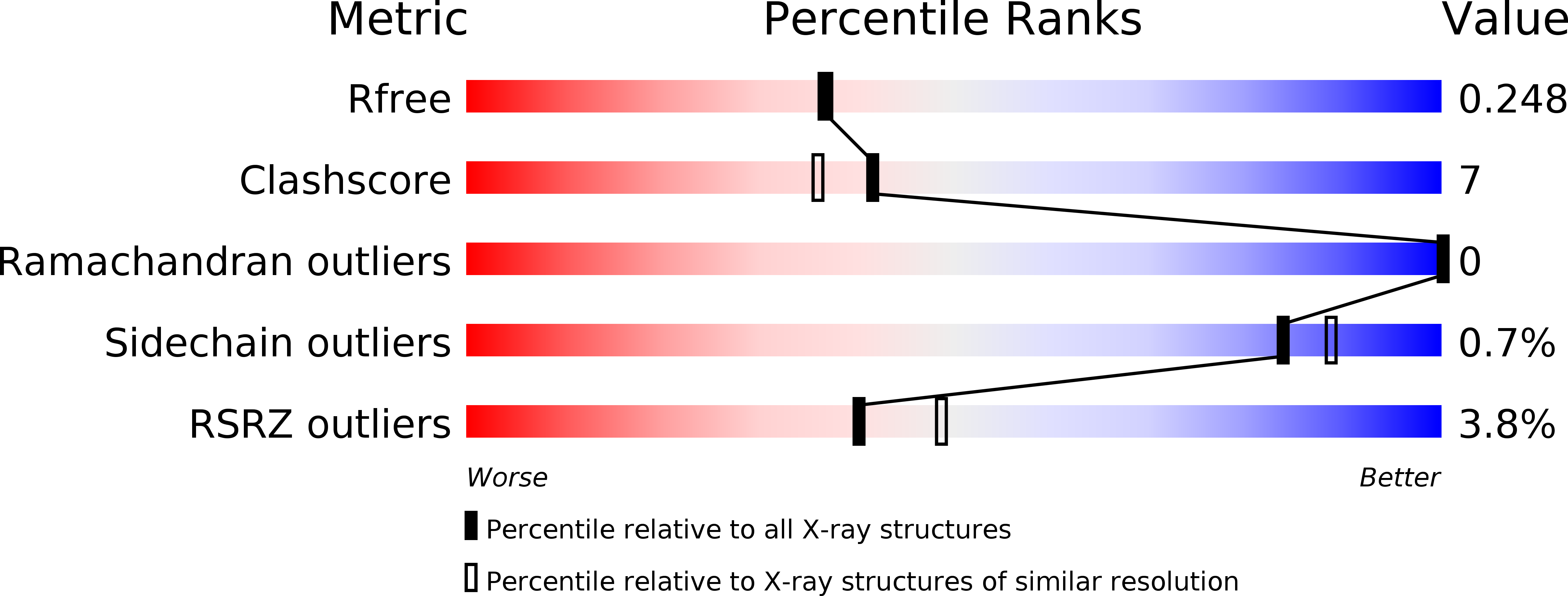
Resolution:
2.15 Å
R-Value Free:
0.24
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 21 21 21