Deposition Date
2019-06-21
Release Date
2019-10-02
Last Version Date
2023-10-11
Entry Detail
PDB ID:
6PFD
Keywords:
Title:
Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)terephthalic acid.
Biological Source:
Source Organism(s):
Cryptosporidium hominis (Taxon ID: 237895)
Expression System(s):
Method Details:
Experimental Method:
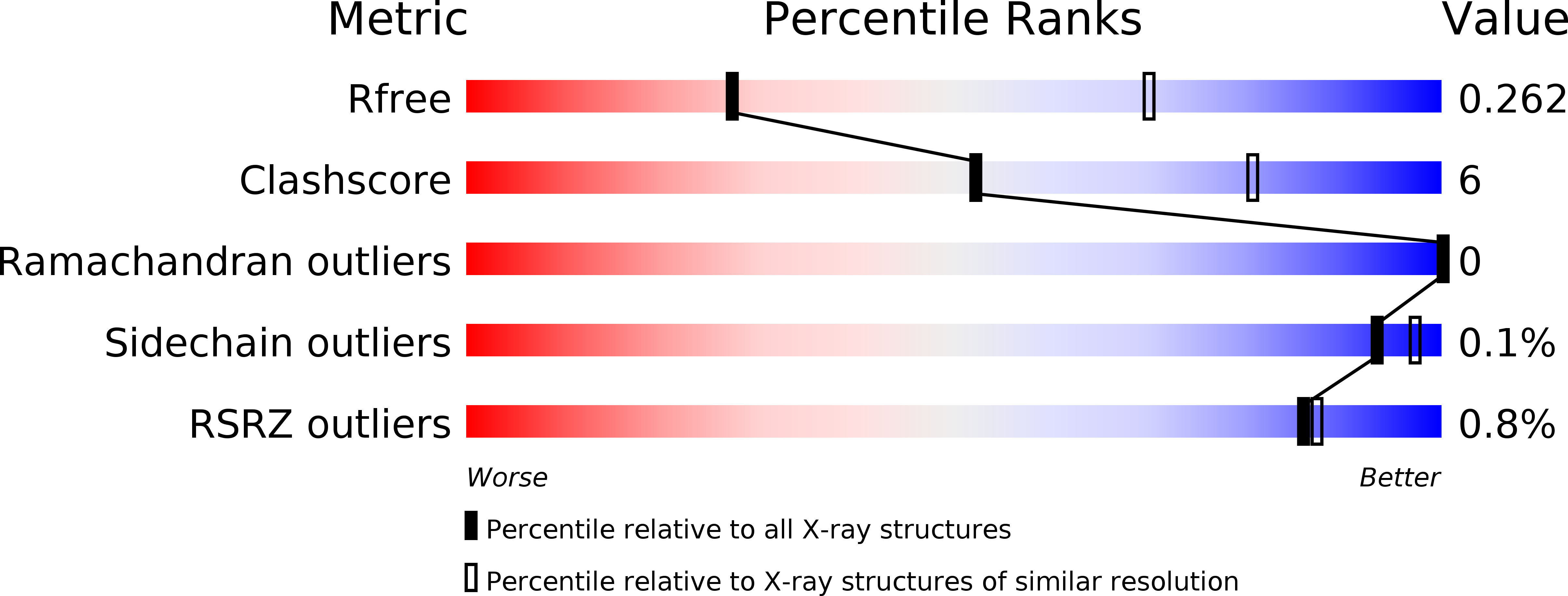
Resolution:
3.32 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.22
Space Group:
C 1 2 1