Deposition Date
2019-06-04
Release Date
2019-08-14
Last Version Date
2024-10-30
Entry Detail
PDB ID:
6P6W
Keywords:
Title:
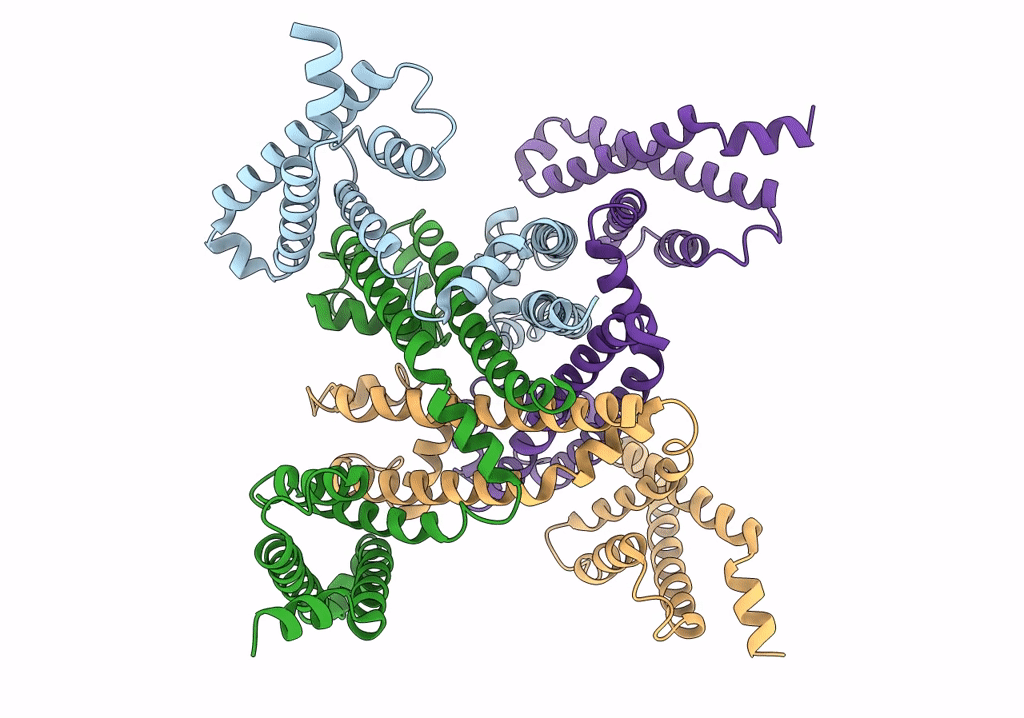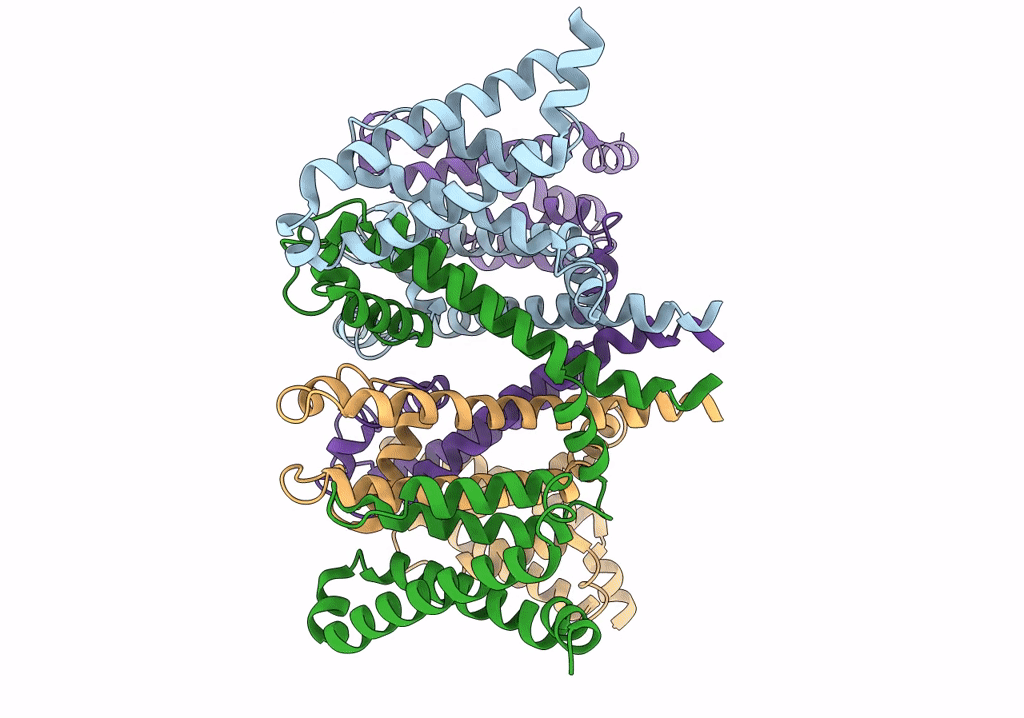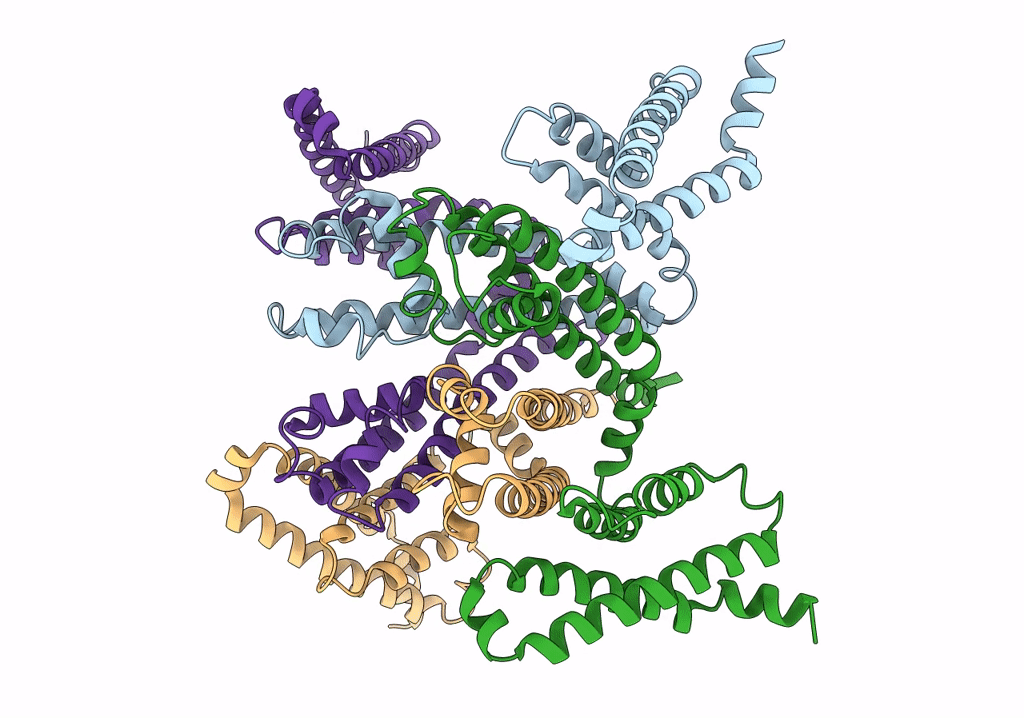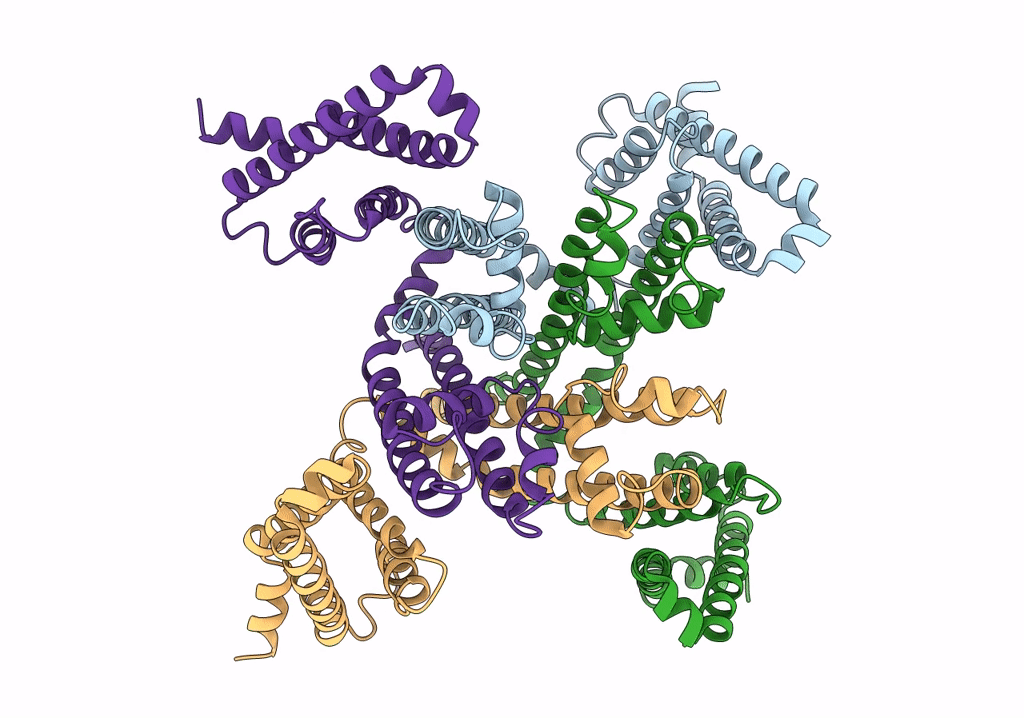
Cryo-EM structure of voltage-gated sodium channel NavAb N49K/L109A/M116V/G94C/Q150C disulfide crosslinked mutant in the resting state
Biological Source:
Source Organism(s):
Escherichia coli K-12 (Taxon ID: 83333)
Arcobacter butzleri (strain RM4018) (Taxon ID: 367737)
Arcobacter butzleri (strain RM4018) (Taxon ID: 367737)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


