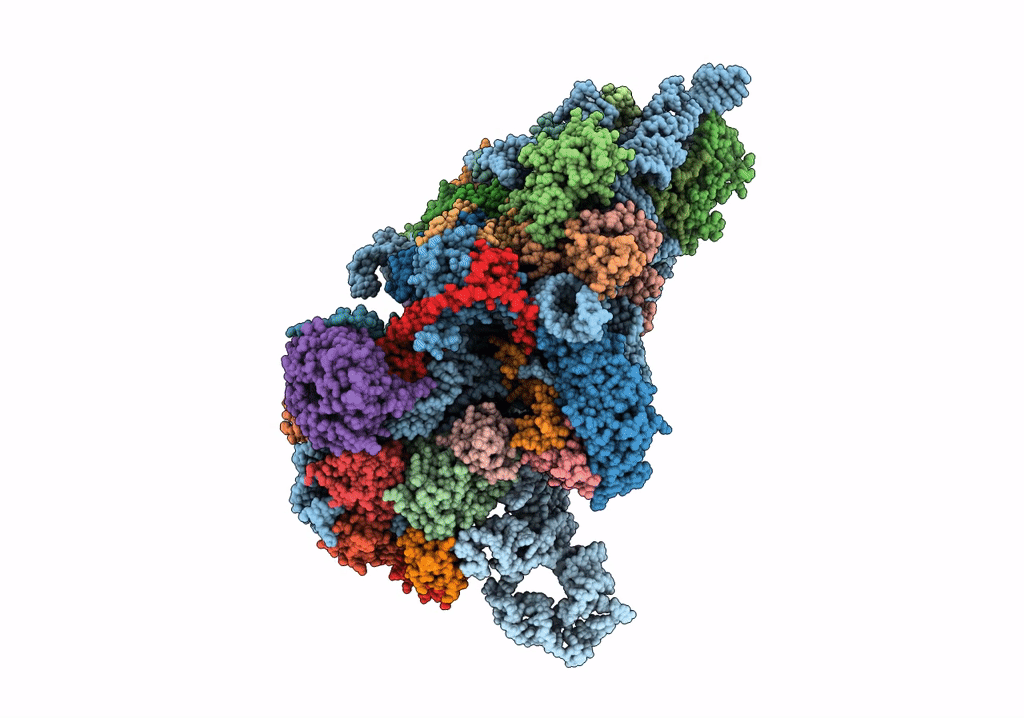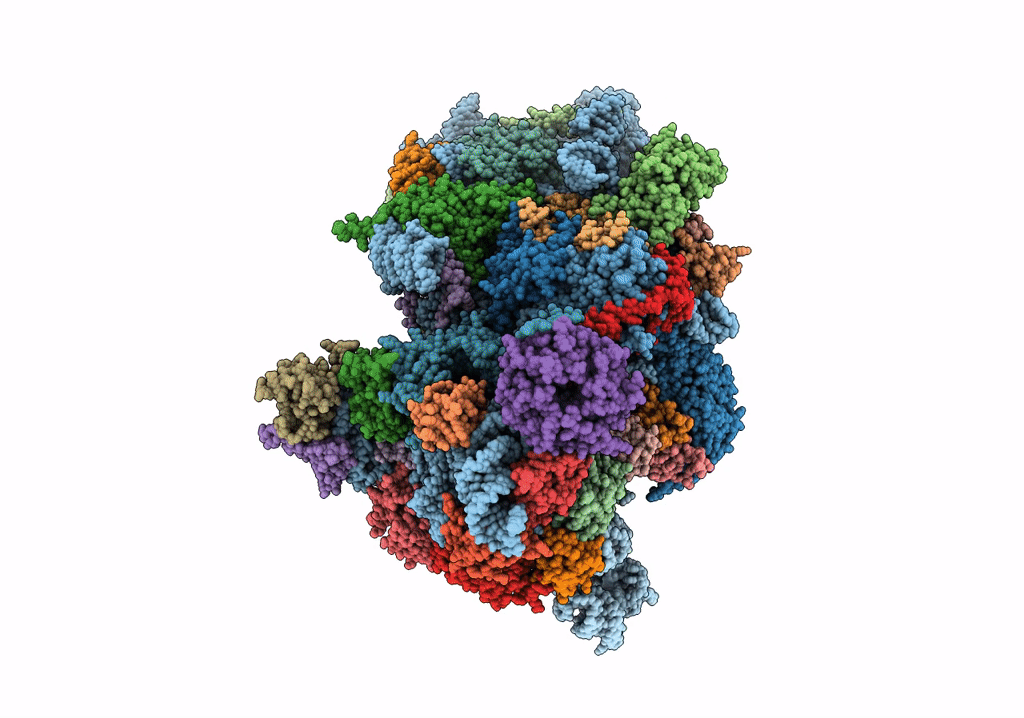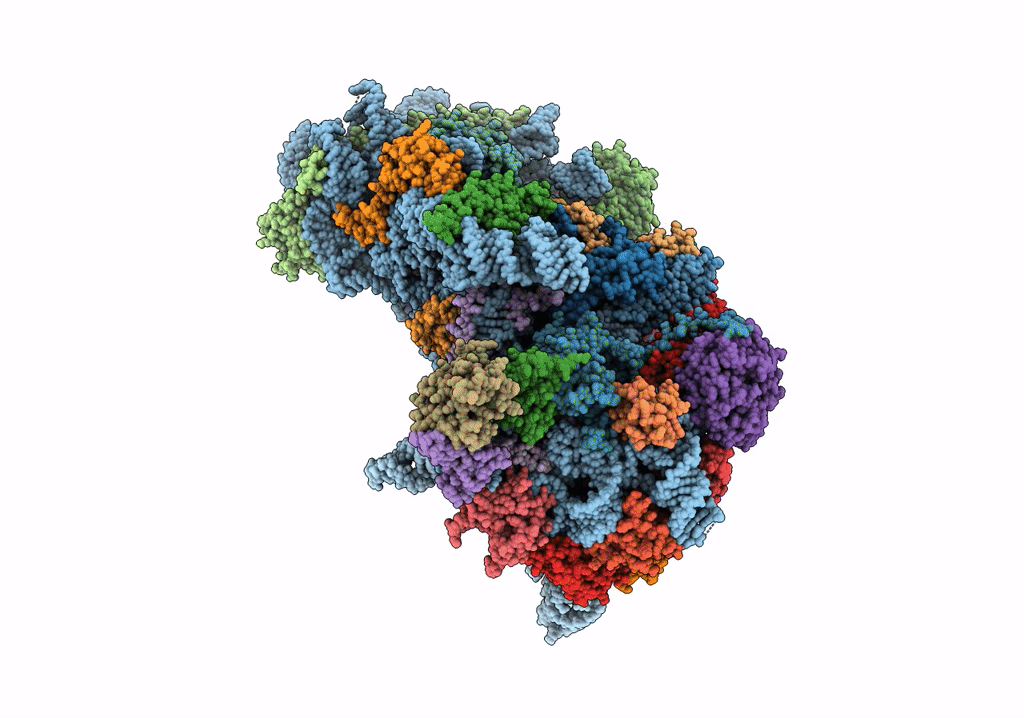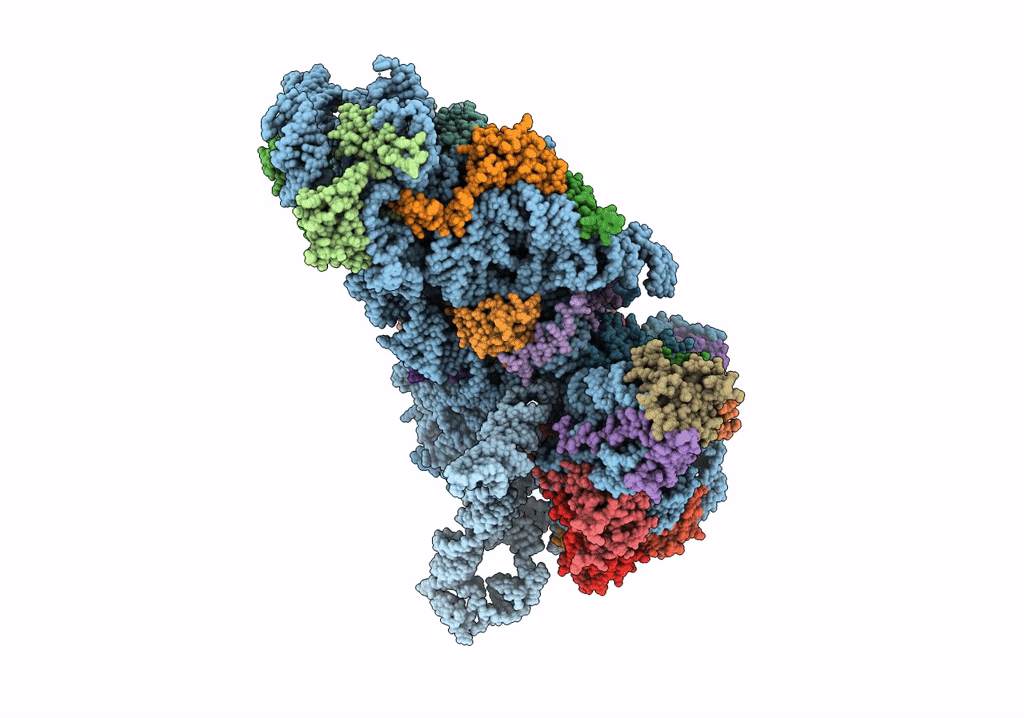
Deposition Date
2019-05-27
Release Date
2019-09-18
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6P4H
Keywords:
Title:
Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2)
Biological Source:
Source Organism(s):
Israeli acute paralysis virus (Taxon ID: 294365)
Oryctolagus cuniculus (Taxon ID: 9986)
Oryctolagus cuniculus (Taxon ID: 9986)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


