Deposition Date
2019-05-13
Release Date
2019-09-11
Last Version Date
2024-10-16
Entry Detail
PDB ID:
6OXL
Keywords:
Title:
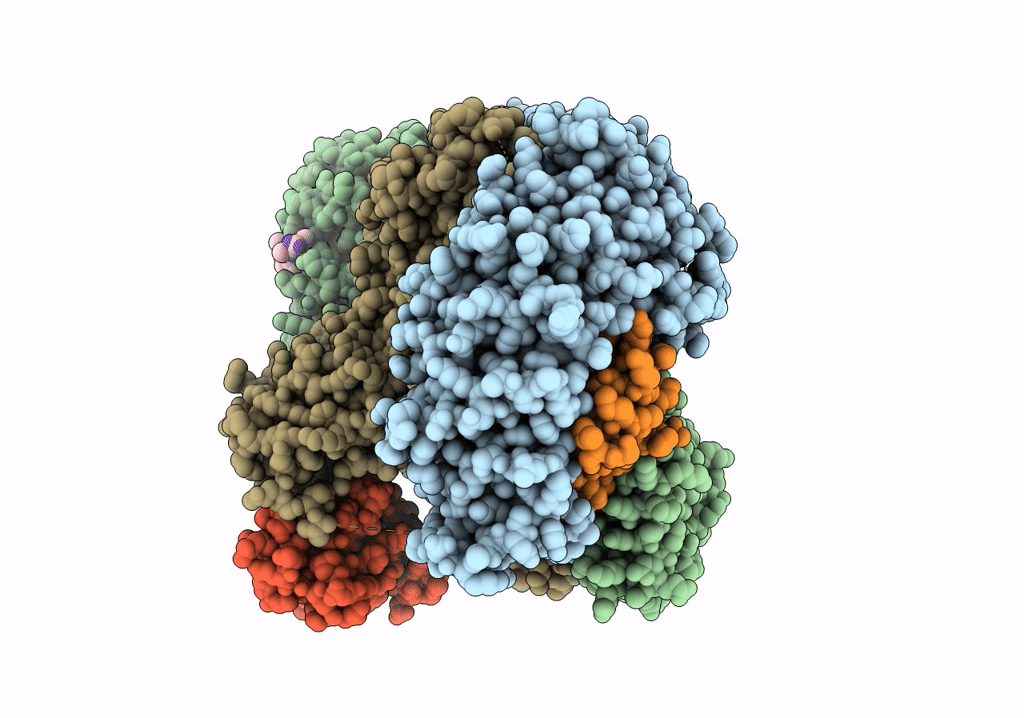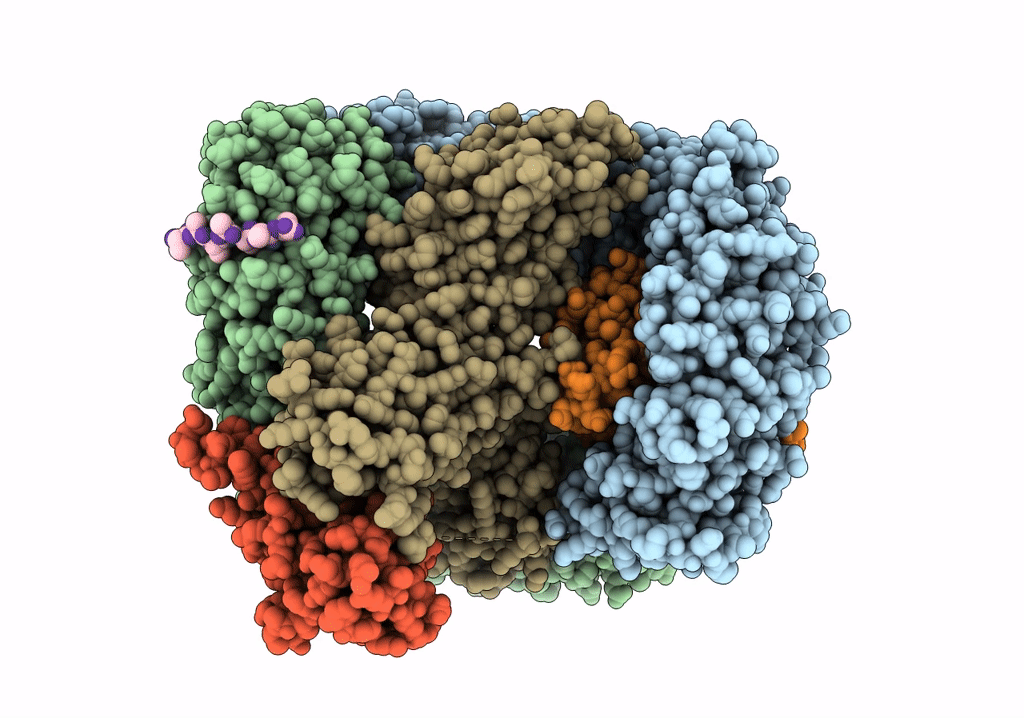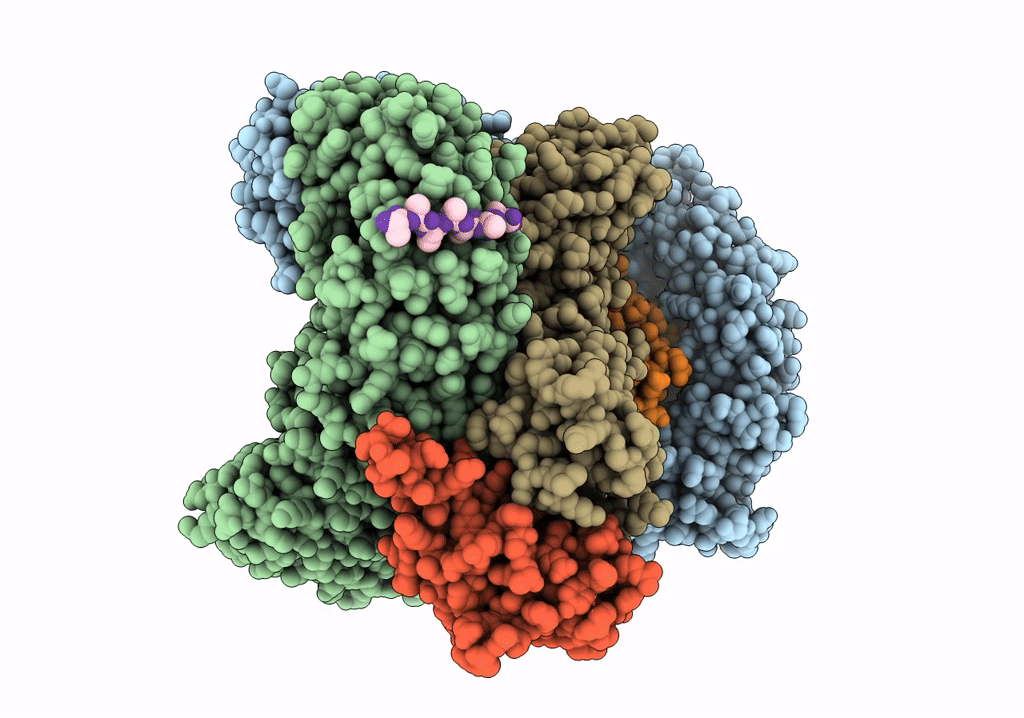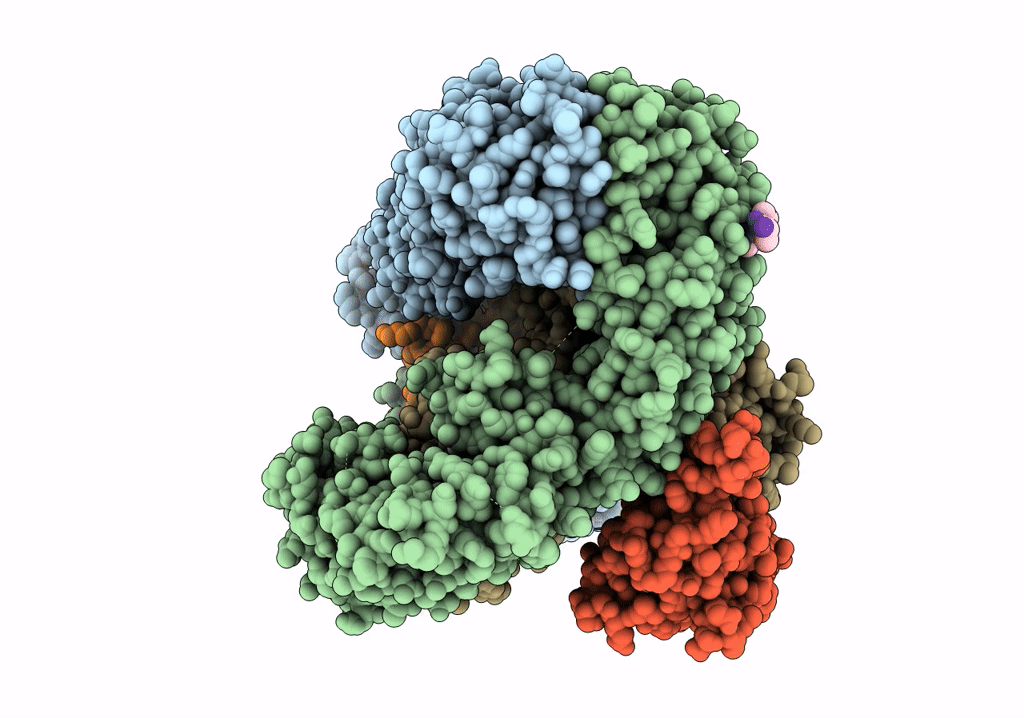
CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 (mu E302K) BOUND TO NECAP IN THE PRESENCE OF SS DNA
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Rattus norvegicus (Taxon ID: 10116)
Rattus norvegicus (Taxon ID: 10116)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


