Deposition Date
2019-05-08
Release Date
2020-03-04
Last Version Date
2024-10-16
Entry Detail
PDB ID:
6OVM
Keywords:
Title:
Crystal Structure of the Pseudomonas capeferrum Anti-sigma Regulator PupR C-terminal Cell-surface Signaling Domain in Complex with the Outer Membrane Transporter PupB N-terminal Signaling Domain (SeMet)
Biological Source:
Source Organism(s):
Pseudomonas capeferrum (Taxon ID: 1495066)
Expression System(s):
Method Details:
Experimental Method:
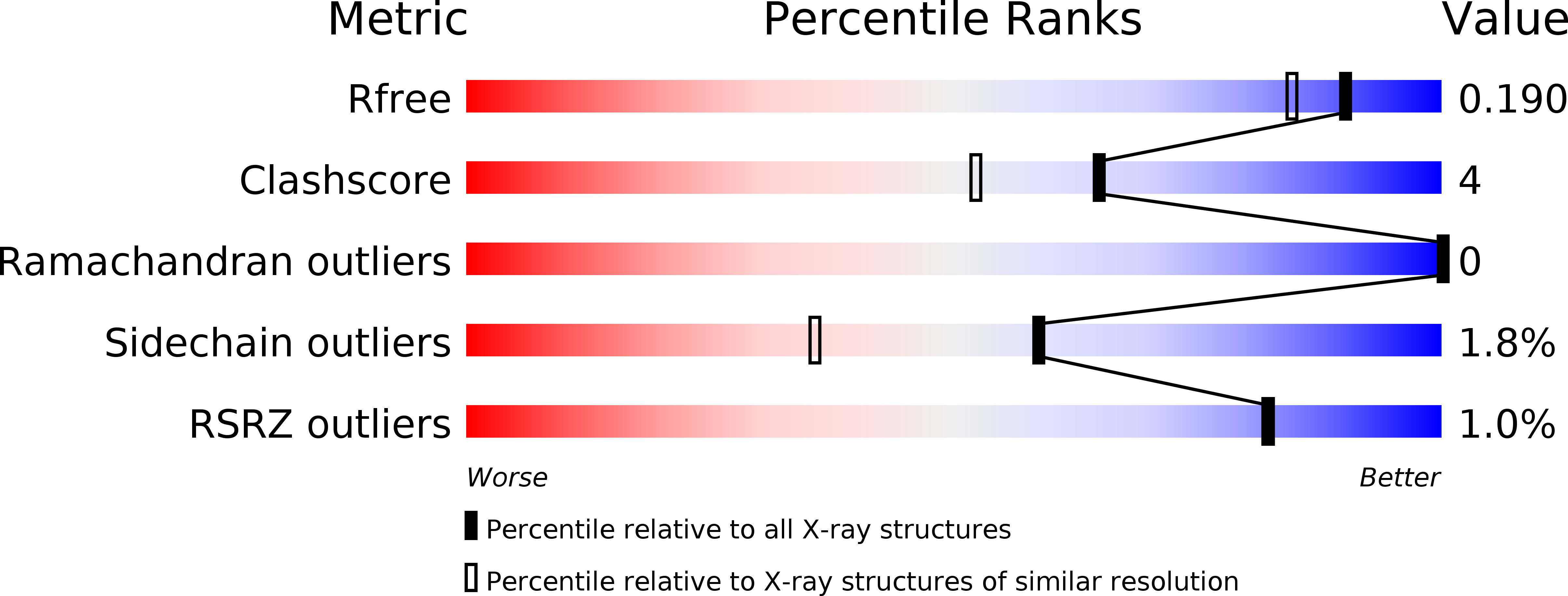
Resolution:
1.60 Å
R-Value Free:
0.18
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
P 21 21 21