Deposition Date
2019-04-09
Release Date
2019-07-31
Last Version Date
2023-10-11
Entry Detail
PDB ID:
6OI9
Keywords:
Title:
Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine
Biological Source:
Source Organism:
Escherichia coli UMNK88 (Taxon ID: 696406)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.06 Å
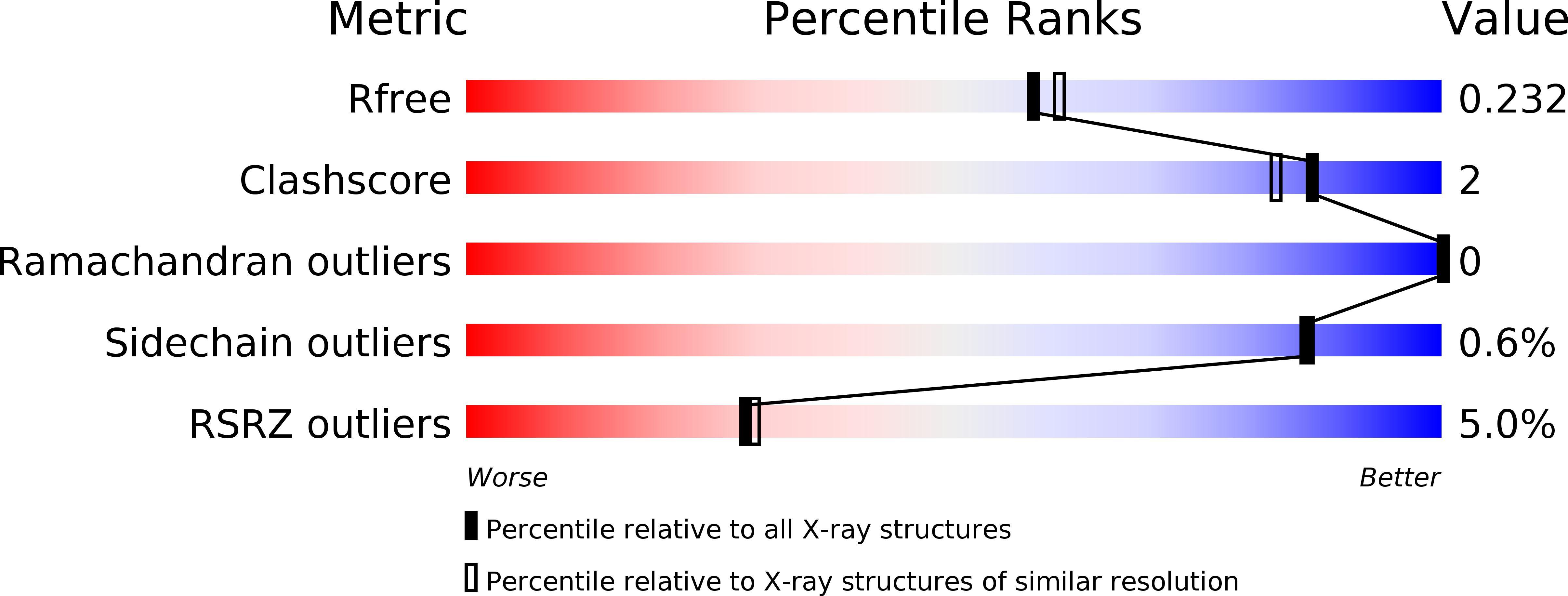
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 21 21 21