Deposition Date
2019-03-26
Release Date
2019-06-19
Last Version Date
2024-11-13
Entry Detail
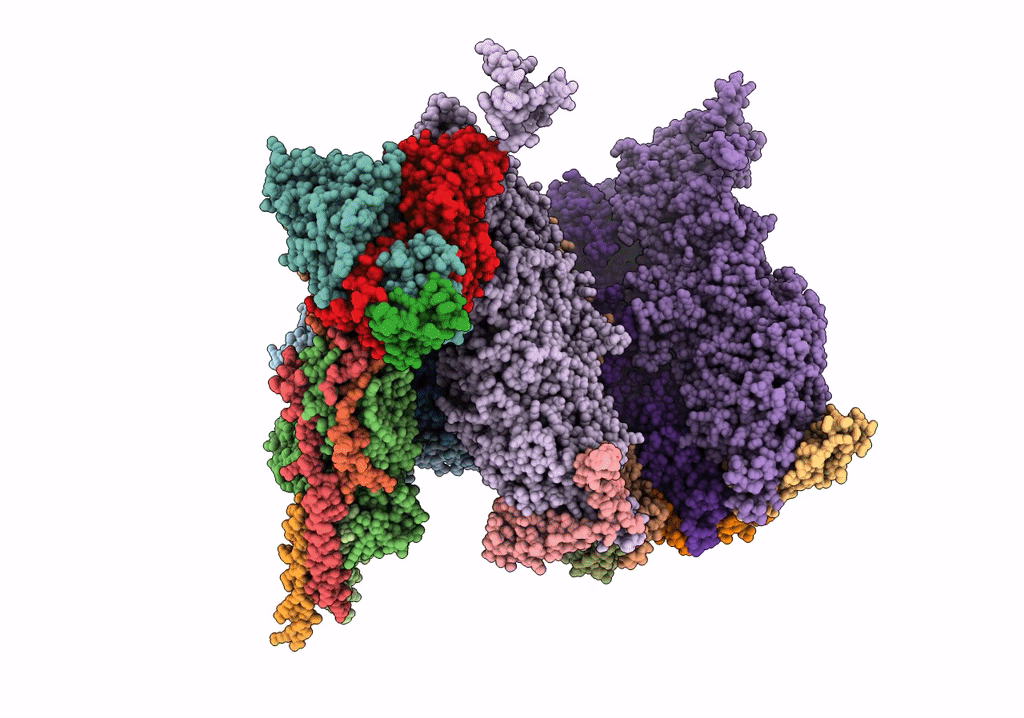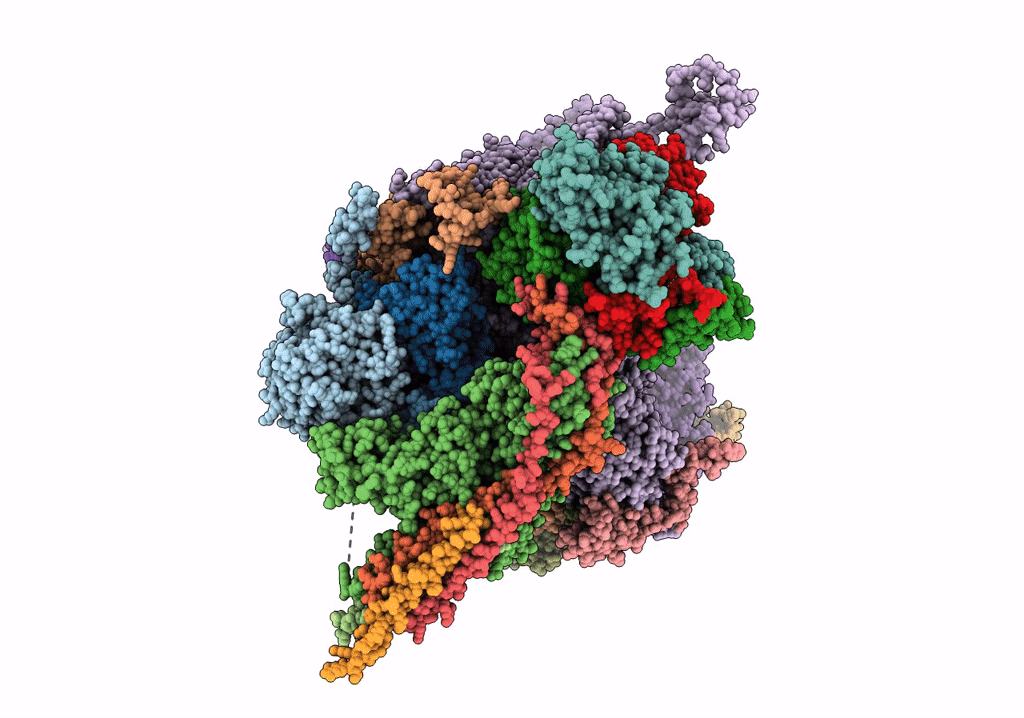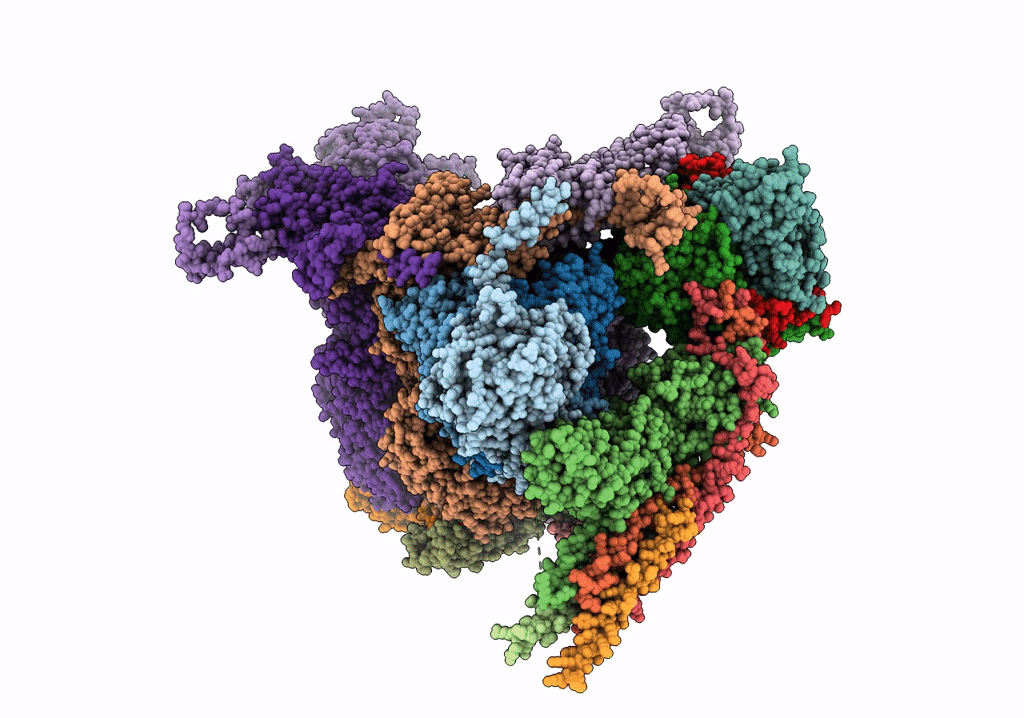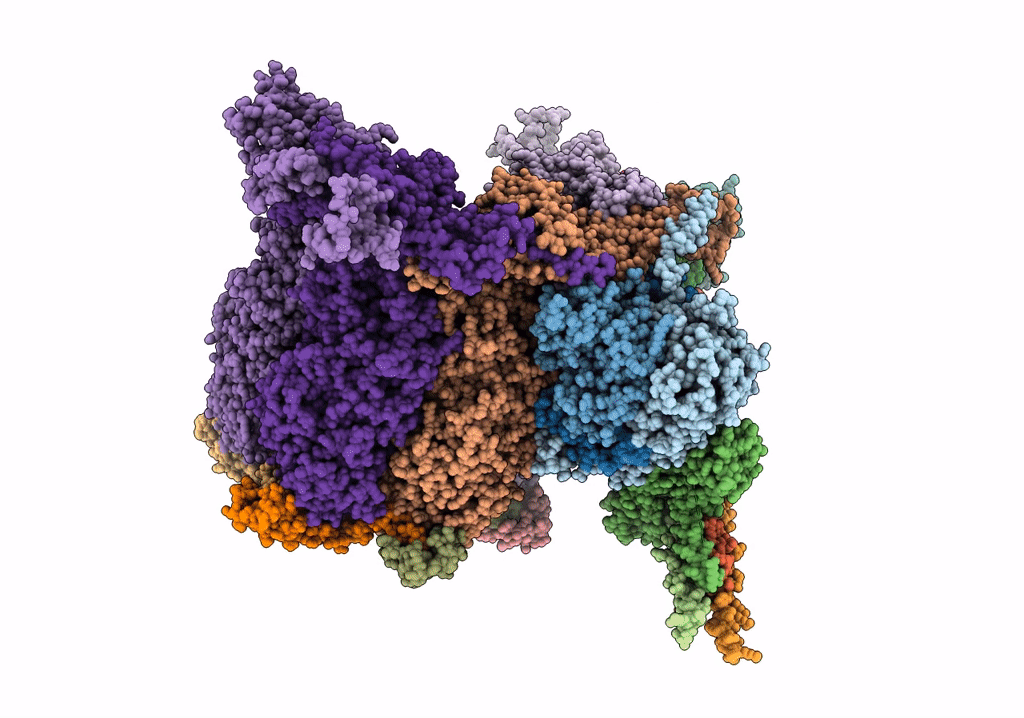
PDB ID:
6ODM
Keywords:
Title:
Herpes simplex virus type 1 (HSV-1) portal vertex-adjacent capsid/CATC, asymmetric unit
Biological Source:
Source Organism(s):
Human herpesvirus 1 strain KOS (Taxon ID: 10306)
Method Details:
Experimental Method:
Resolution:
4.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE