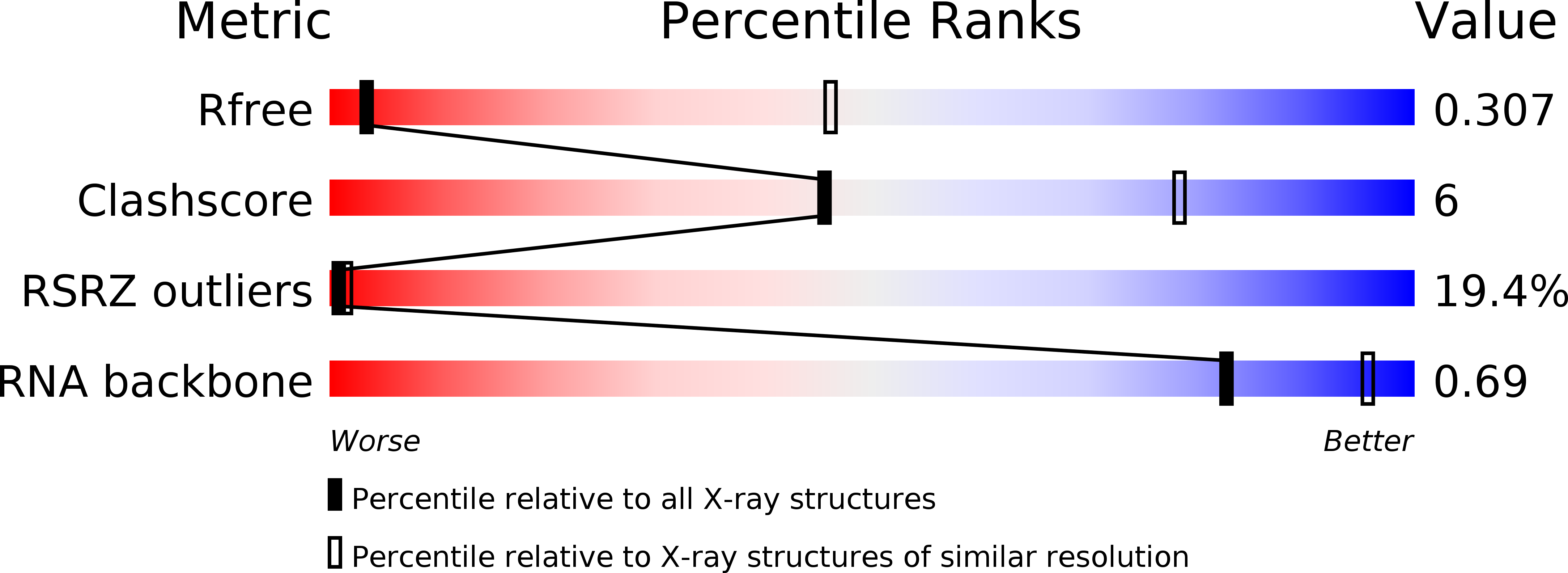Deposition Date
2019-03-26
Release Date
2019-07-17
Last Version Date
2023-10-11
Entry Detail
PDB ID:
6OD9
Keywords:
Title:
Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser
Biological Source:
Source Organism(s):
Fusobacterium ulcerans (Taxon ID: 861)
Method Details:
Experimental Method:
Resolution:
4.10 Å
R-Value Free:
0.30
R-Value Work:
0.25
R-Value Observed:
0.26
Space Group:
P 32 2 1