Deposition Date
2019-03-07
Release Date
2019-07-31
Last Version Date
2024-03-20
Entry Detail
PDB ID:
6O7I
Keywords:
Title:
Cryo-EM structure of Csm-crRNA-target RNA ternary bigger complex in complex with cA4 in type III-A CRISPR-Cas system
Biological Source:
Source Organism:
Thermococcus onnurineus (strain NA1) (Taxon ID: 523850)
Thermococcus onnurineus (Taxon ID: 342948)
synthetic construct (Taxon ID: 32630)
Thermococcus onnurineus (Taxon ID: 342948)
synthetic construct (Taxon ID: 32630)
Host Organism:
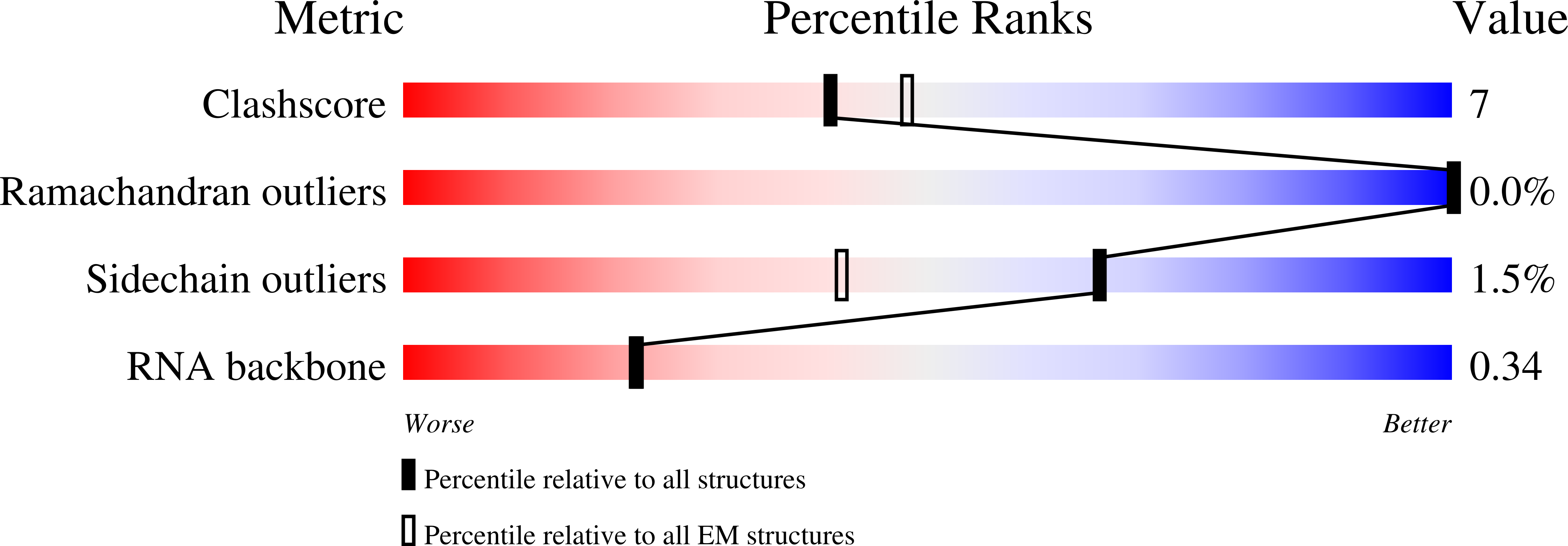
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE