Deposition Date
2018-11-19
Release Date
2020-05-27
Last Version Date
2024-11-06
Entry Detail
PDB ID:
6N4C
Keywords:
Title:
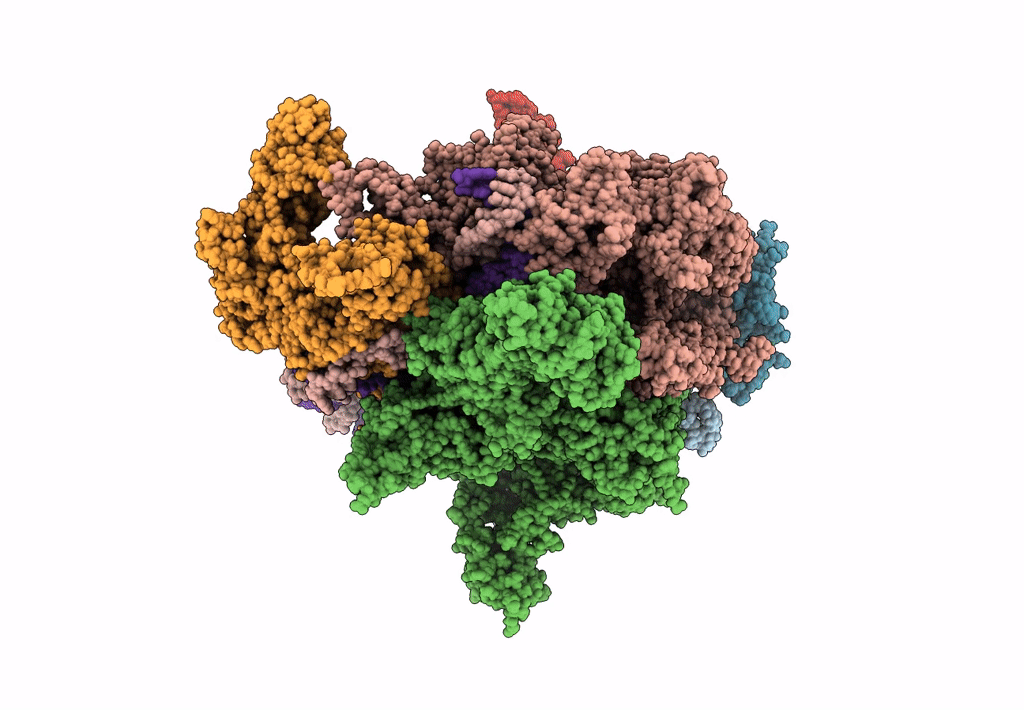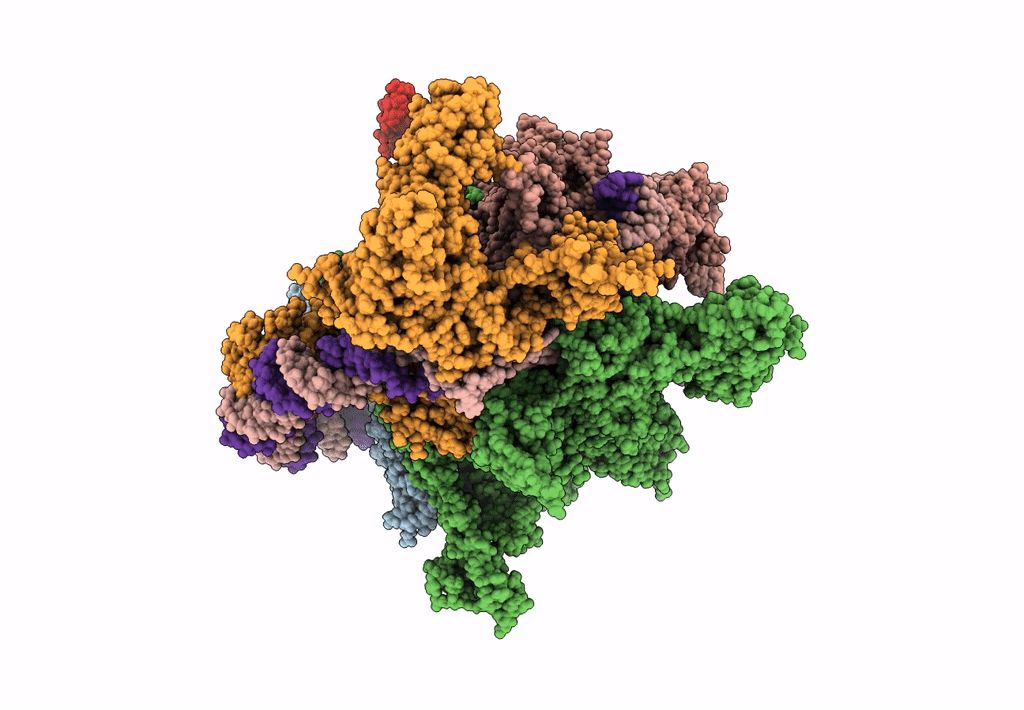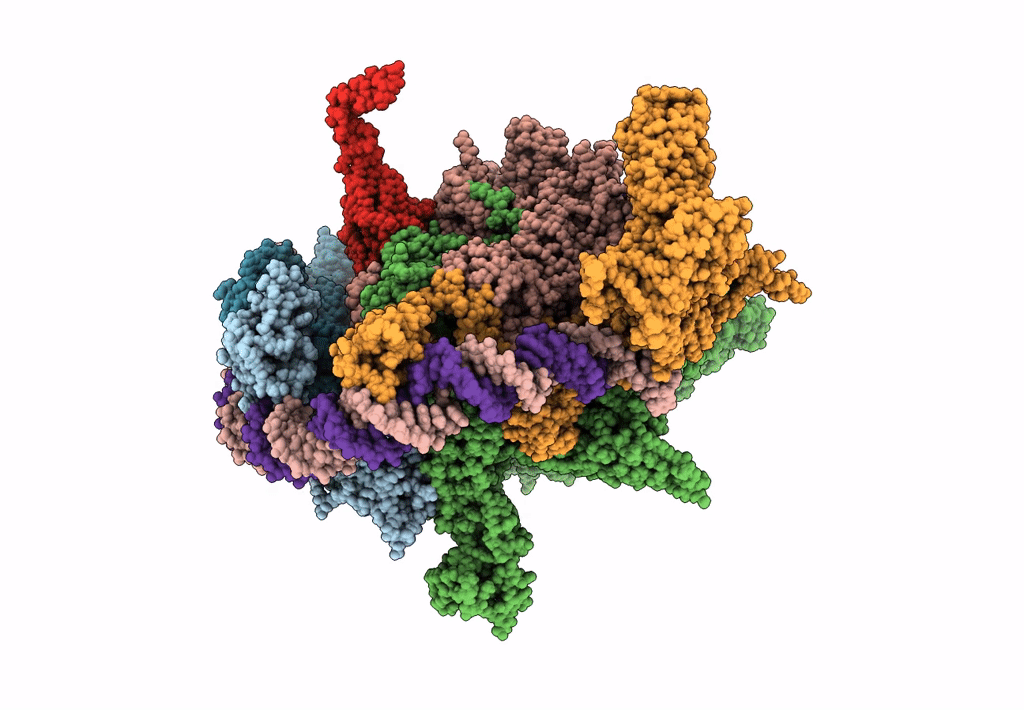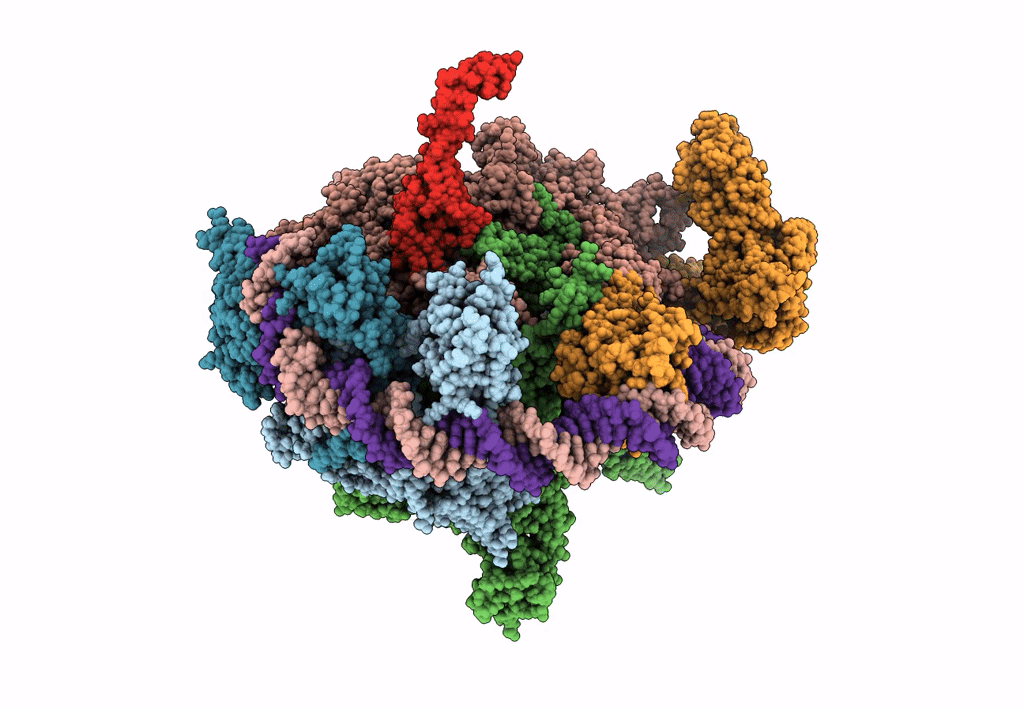
EM structure of the DNA wrapping in bacterial open transcription initiation complex
Biological Source:
Source Organism:
Enterobacteria phage lambda (Taxon ID: 10710)
Escherichia coli K-12 (Taxon ID: 83333)
Escherichia coli K-12 (Taxon ID: 83333)
Method Details:
Experimental Method:
Resolution:
17.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


