Deposition Date
2018-10-05
Release Date
2019-04-24
Last Version Date
2023-10-11
Entry Detail
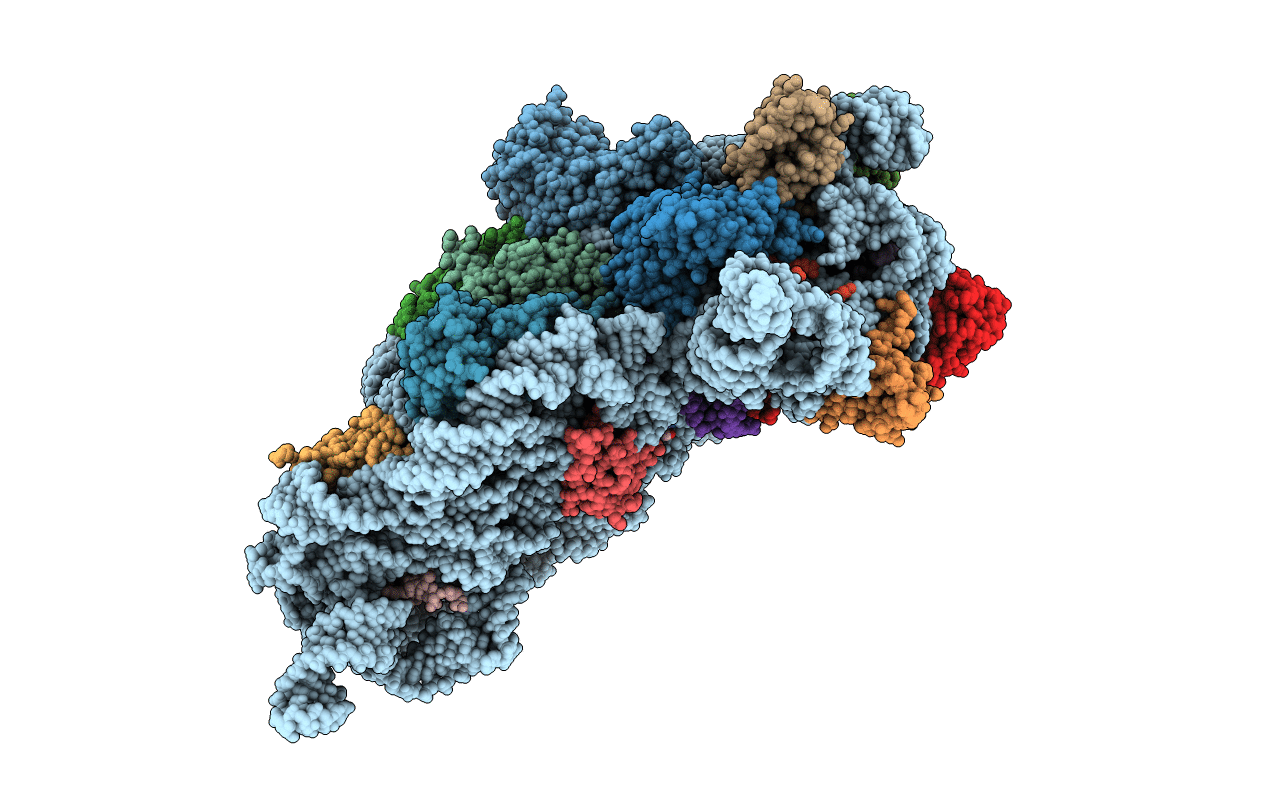
PDB ID:
6MPF
Keywords:
Title:
Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGC-codon in the A-site and paromomycin
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Thermus thermophilus HB8 (strain HB8 / ATCC 27634 / DSM 579) (Taxon ID: 300852)
Thermus thermophilus HB8 (strain HB8 / ATCC 27634 / DSM 579) (Taxon ID: 300852)
Method Details:
Experimental Method:
Resolution:
3.33 Å
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 41 21 2


