Deposition Date
2018-09-18
Release Date
2019-05-29
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6MI1
Keywords:
Title:
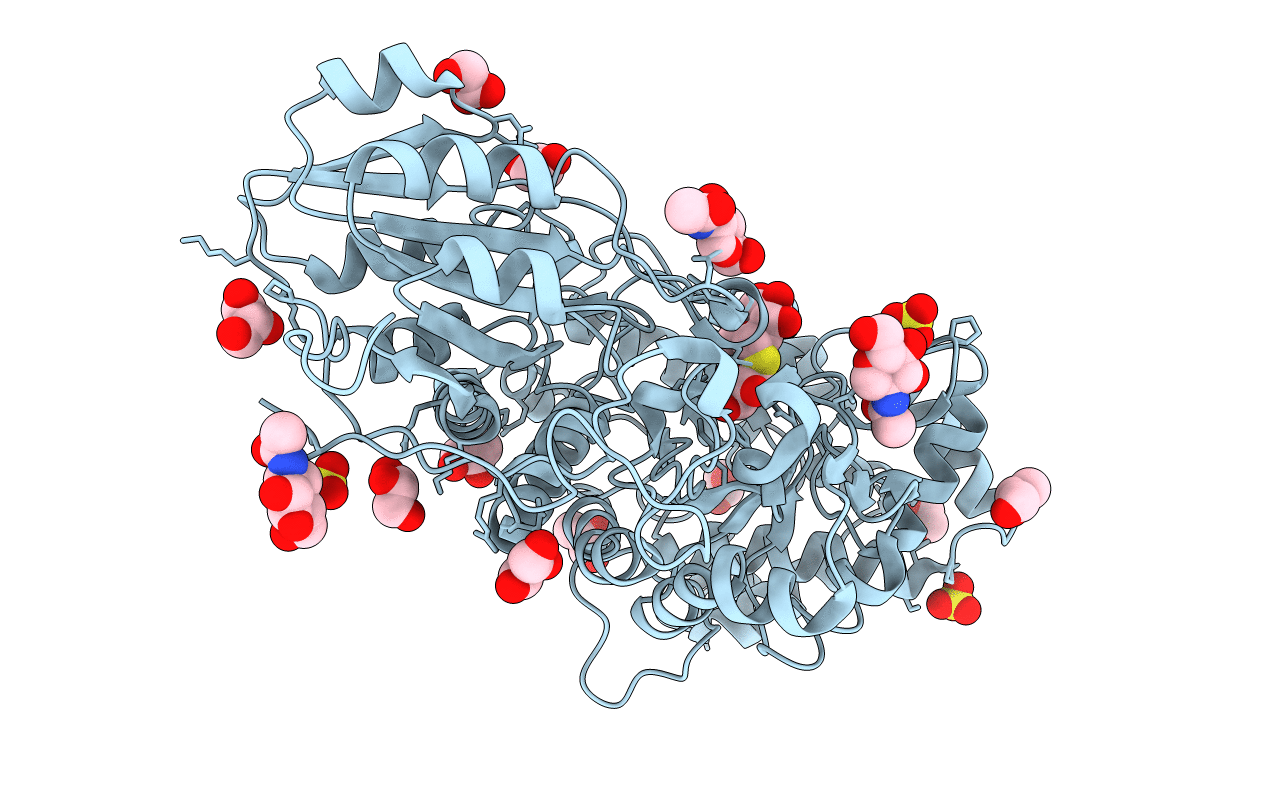
CRYSTAL STRUCTURE ANALYSIS OF THE VARIANT PLANT EXOHYDROLASE ARG158ALA-GLU161ALA IN COMPLEX WITH METHYL 6-THIO-BETA-GENTIOBIOSIDE
Biological Source:
Source Organism(s):
Hordeum vulgare subsp. vulgare (Taxon ID: 112509)
Expression System(s):
Method Details:
Experimental Method:
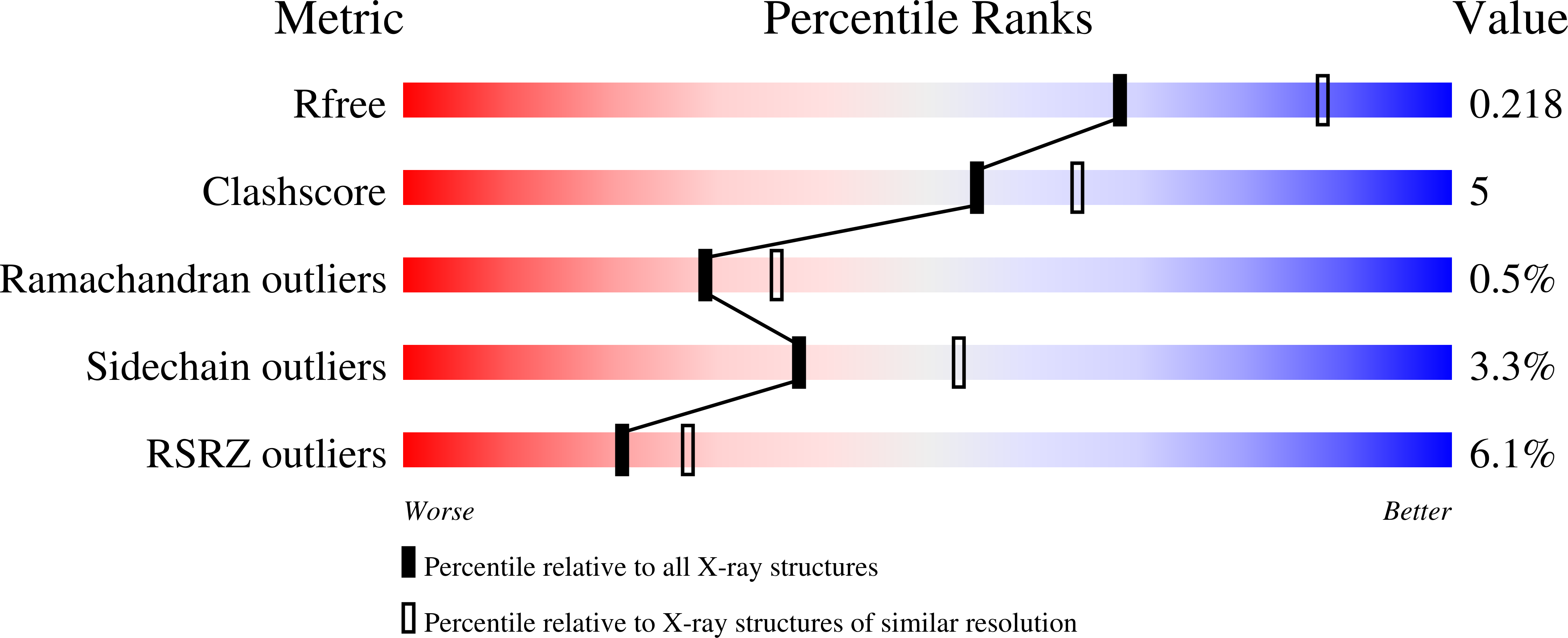
Resolution:
2.30 Å
R-Value Free:
0.21
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 43 21 2