Deposition Date
2018-09-04
Release Date
2018-09-19
Last Version Date
2024-03-13
Entry Detail
PDB ID:
6MDO
Keywords:
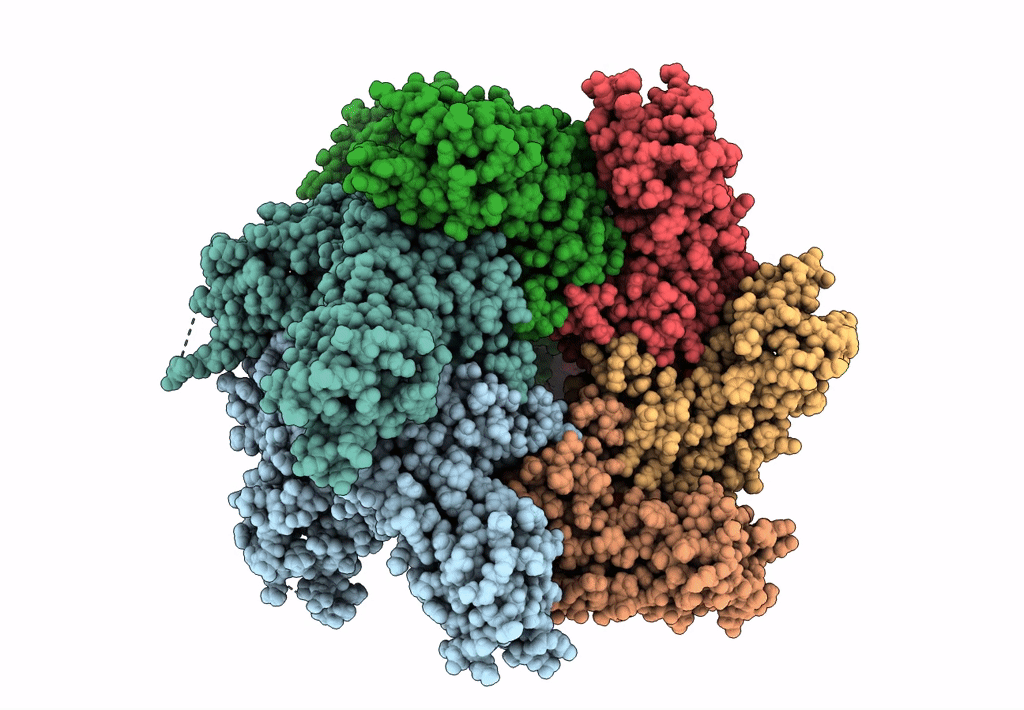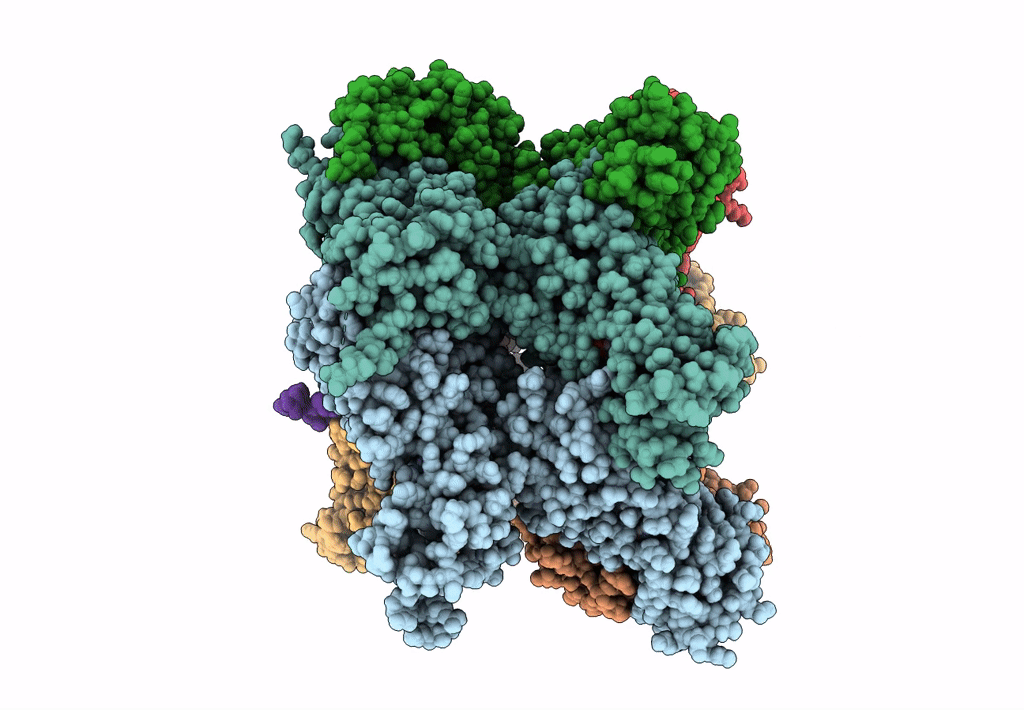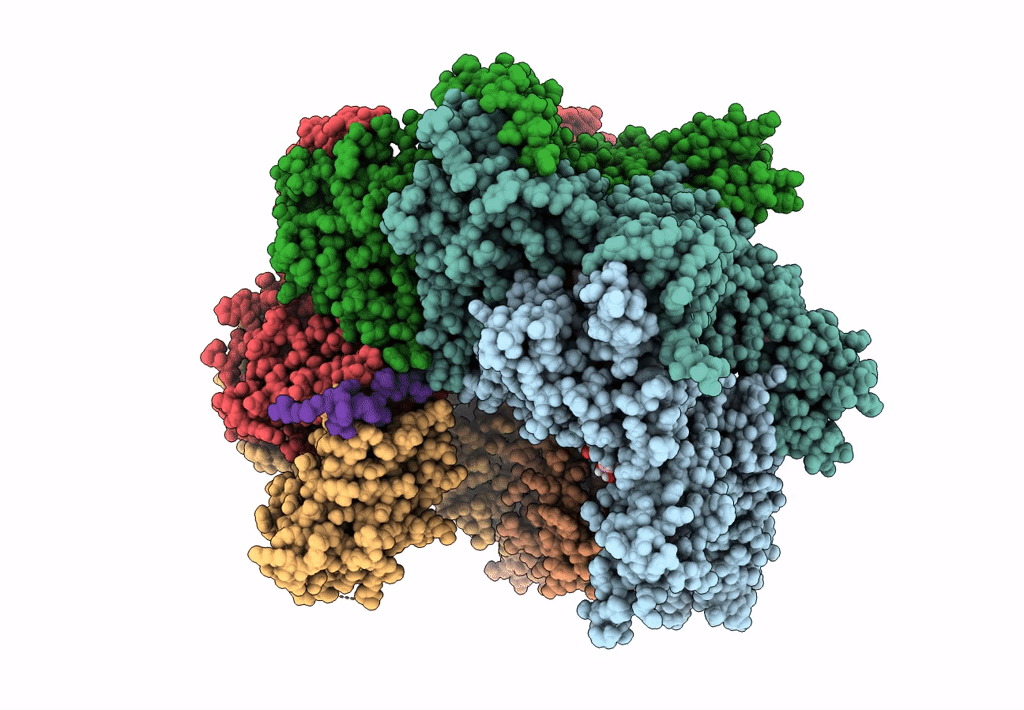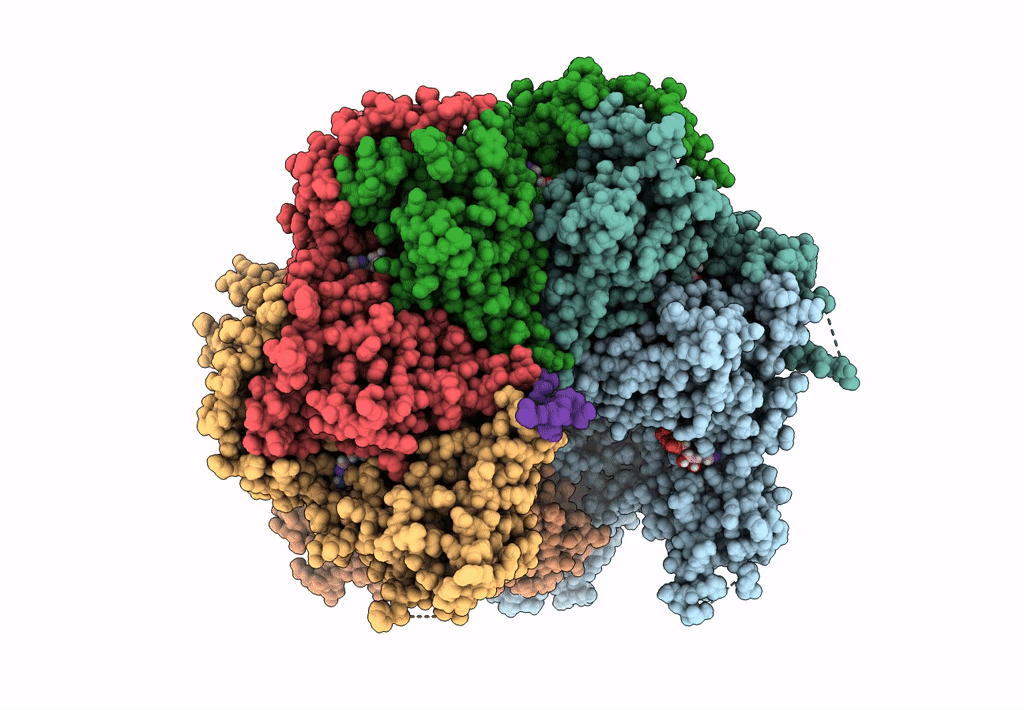
Title:
The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1)
Biological Source:
Source Organism(s):
Cricetulus griseus (Taxon ID: 10029)
Rattus norvegicus (Taxon ID: 10116)
Rattus norvegicus (Taxon ID: 10116)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


