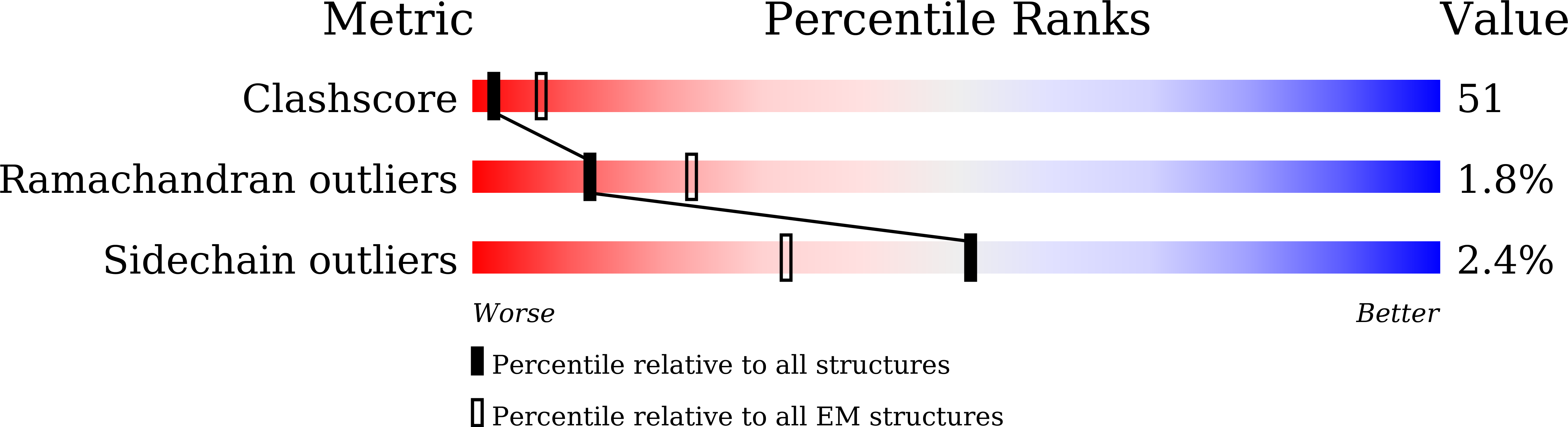Deposition Date
2019-10-25
Release Date
2020-02-12
Last Version Date
2024-11-20
Entry Detail
PDB ID:
6L62
Keywords:
Title:
Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Porcine circovirus 2 (Taxon ID: 85708)
Porcine circovirus 2 (Taxon ID: 85708)
Method Details:
Experimental Method:
Resolution:
7.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE