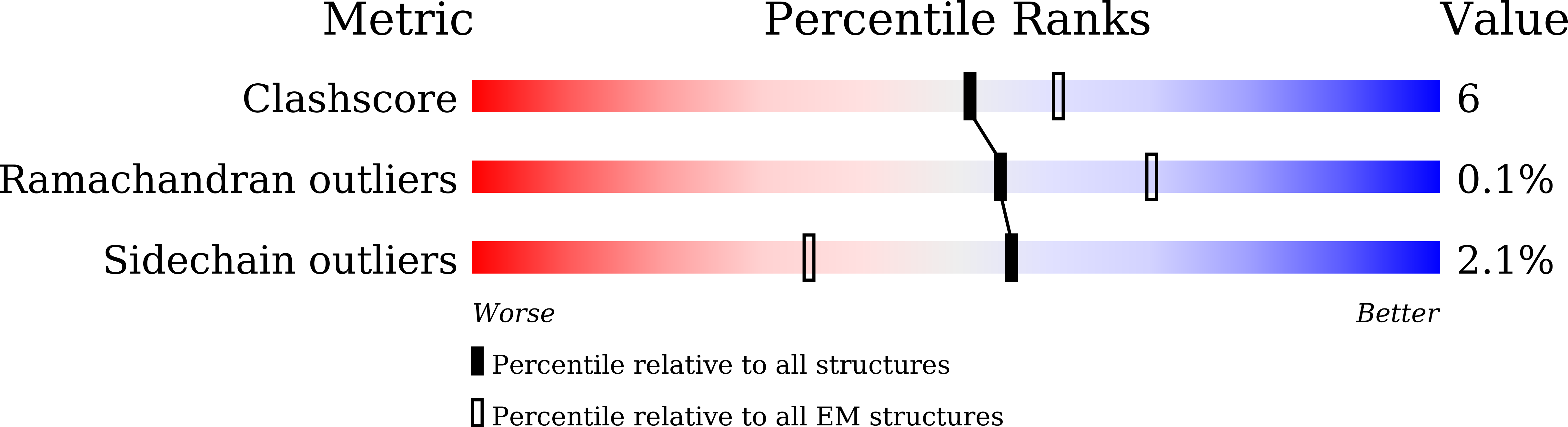
Deposition Date
2019-08-01
Release Date
2020-01-15
Last Version Date
2024-03-27
Entry Detail
PDB ID:
6KMX
Keywords:
Title:
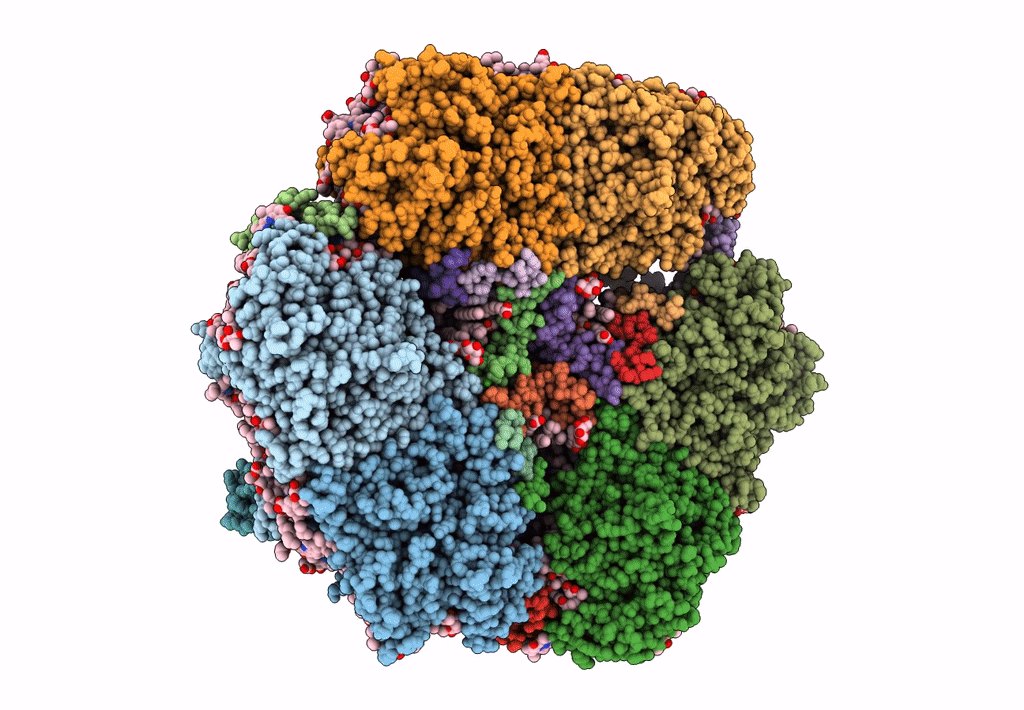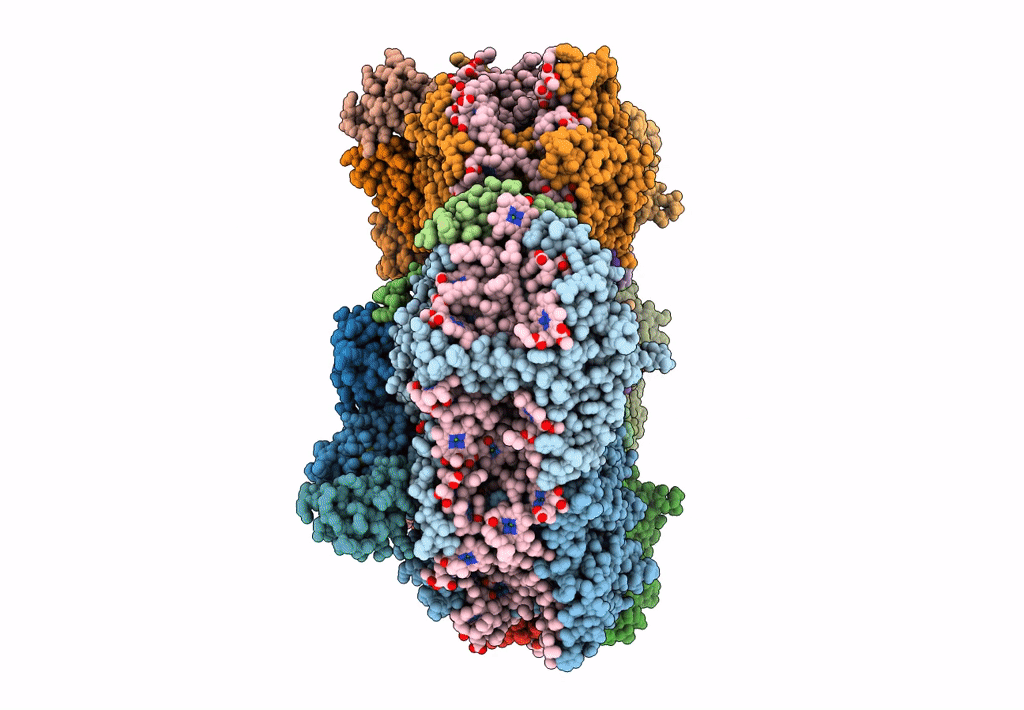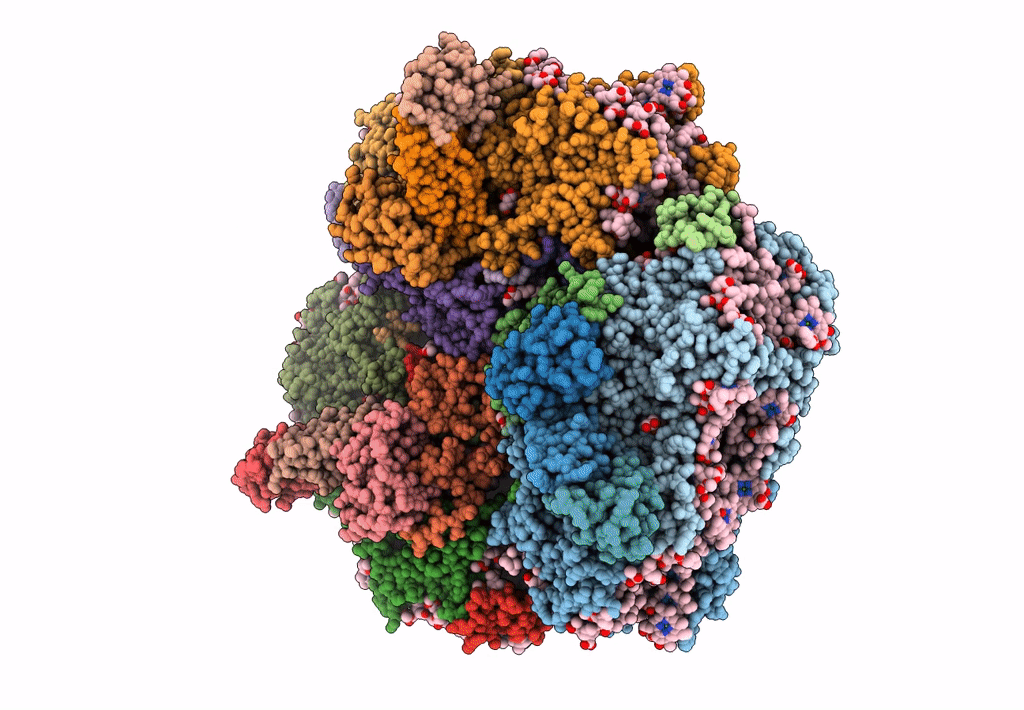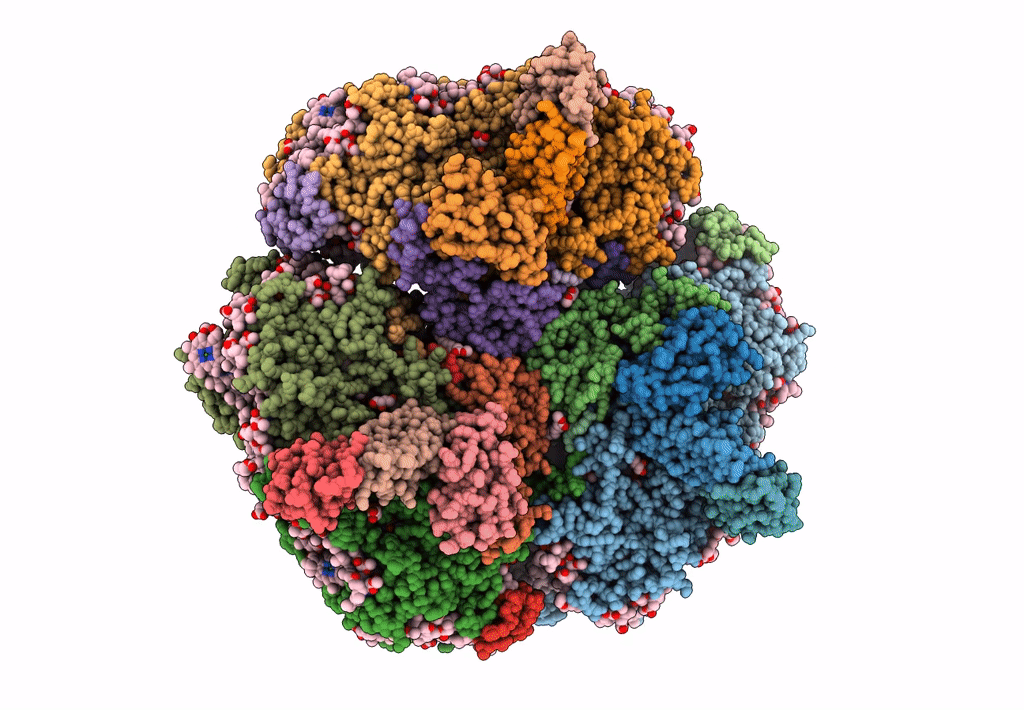
Structure of PSI from H. hongdechloris grown under far-red light condition
Biological Source:
Source Organism(s):
Halomicronema hongdechloris C2206 (Taxon ID: 1641165)
Method Details:
Experimental Method:
Resolution:
2.41 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE