Deposition Date
2019-07-23
Release Date
2019-11-13
Last Version Date
2024-10-16
Entry Detail
PDB ID:
6KJU
Keywords:
Title:
Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors
Biological Source:
Source Organism:
Vibrio cholerae (Taxon ID: 666)
Host Organism:
Method Details:
Experimental Method:
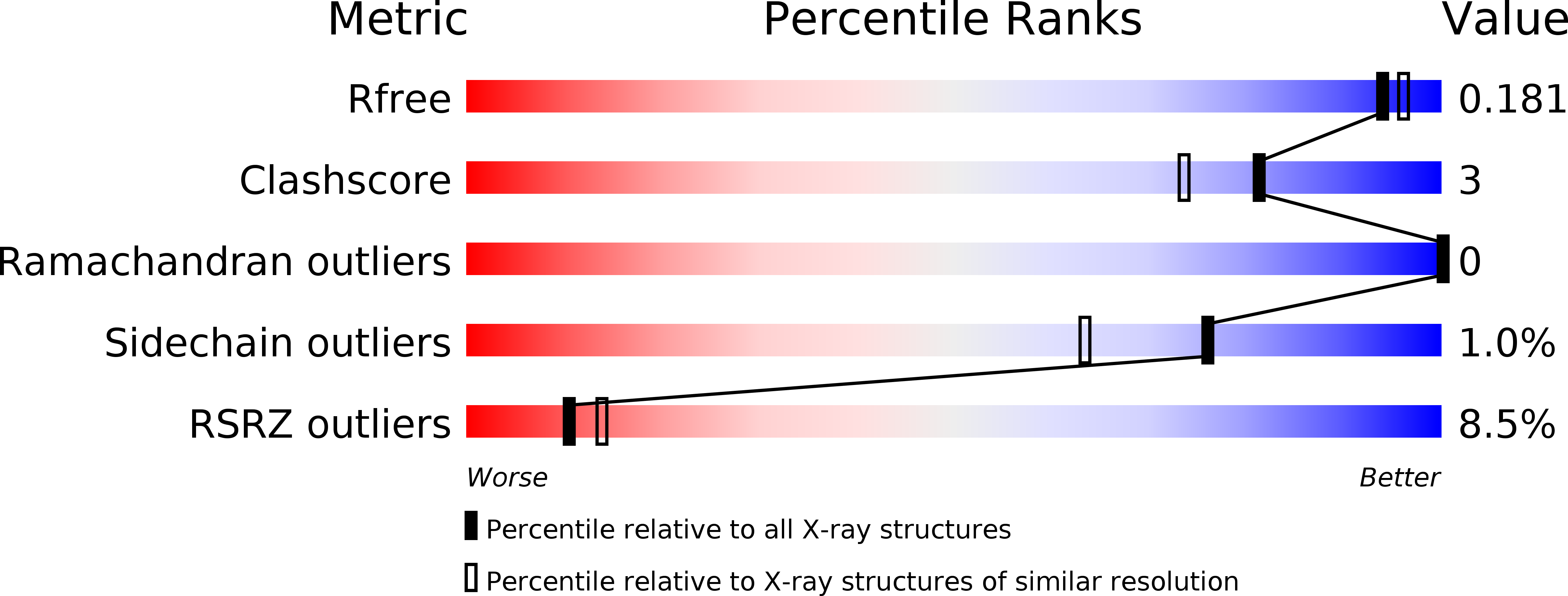
Resolution:
1.75 Å
R-Value Free:
0.19
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 21 21 21