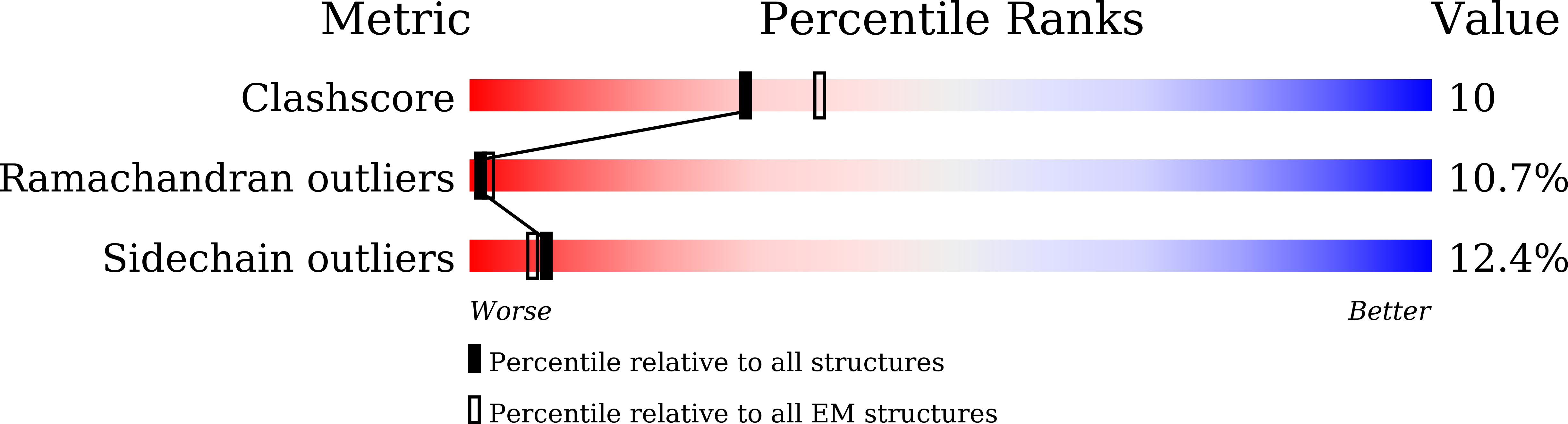
Deposition Date
2019-07-19
Release Date
2020-03-04
Last Version Date
2024-03-27
Entry Detail
PDB ID:
6KIQ
Keywords:
Title:
Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae S288c (Taxon ID: 559292)
Sus scrofa (Taxon ID: 9823)
Sus scrofa (Taxon ID: 9823)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.62 Å
Aggregation State:
FILAMENT
Reconstruction Method:
HELICAL