Deposition Date
2019-05-29
Release Date
2019-09-25
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6K5J
Keywords:
Title:
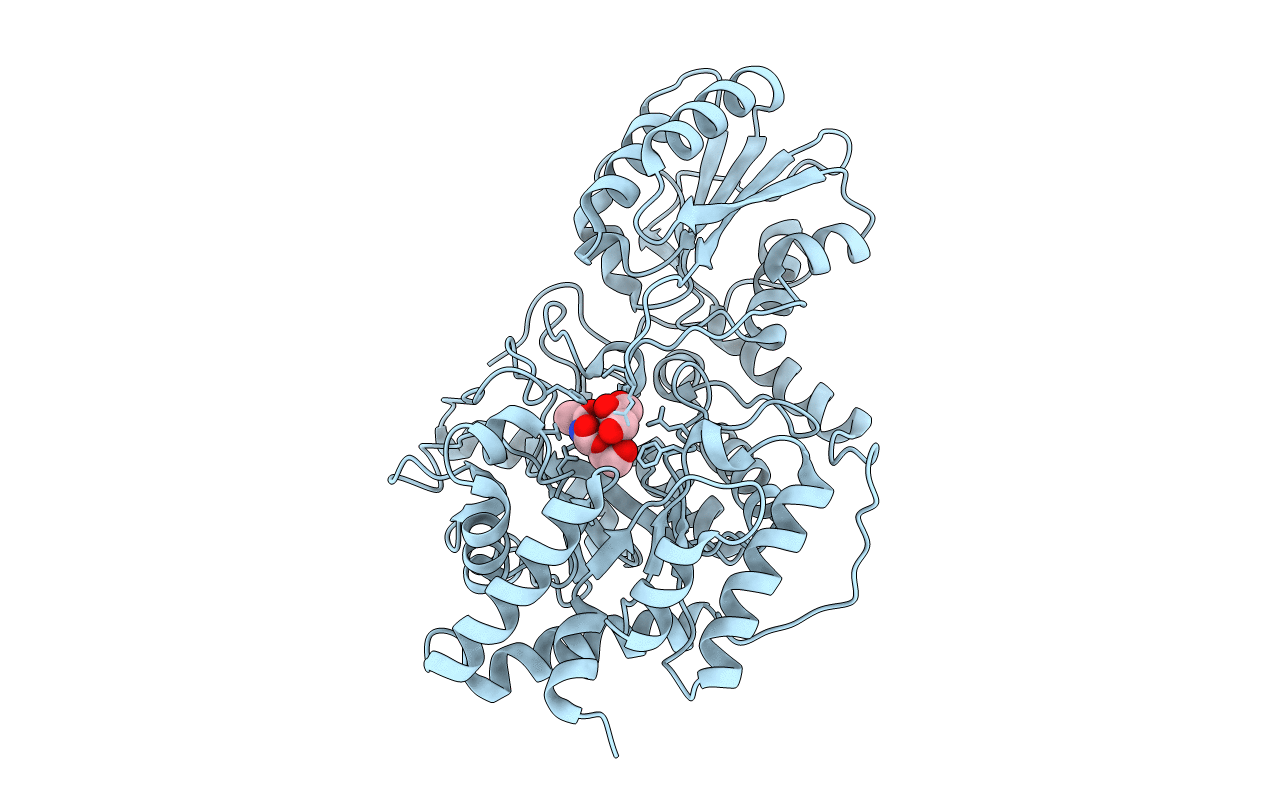
Structure of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7
Biological Source:
Source Organism:
Paenibacillaceae (Taxon ID: 186822)
Host Organism:
Method Details:
Experimental Method:
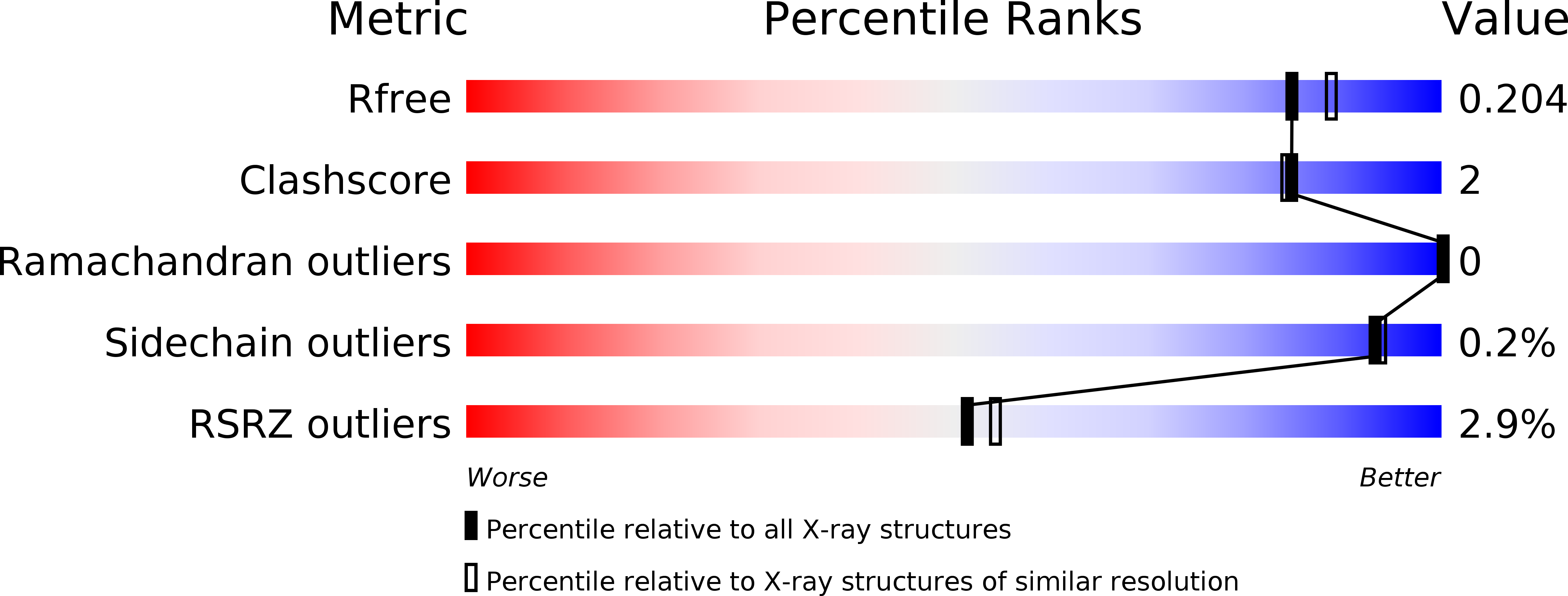
Resolution:
1.90 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 43 21 2