Deposition Date
2019-05-05
Release Date
2019-06-19
Last Version Date
2024-03-27
Entry Detail
PDB ID:
6K0B
Keywords:
Title:
cryo-EM structure of archaeal Ribonuclease P with mature tRNA
Biological Source:
Source Organism(s):
Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) (Taxon ID: 243232)
Escherichia coli (Taxon ID: 562)
Methanocaldococcus jannaschii (Taxon ID: 2190)
Escherichia coli (Taxon ID: 562)
Methanocaldococcus jannaschii (Taxon ID: 2190)
Expression System(s):
Method Details:
Experimental Method:
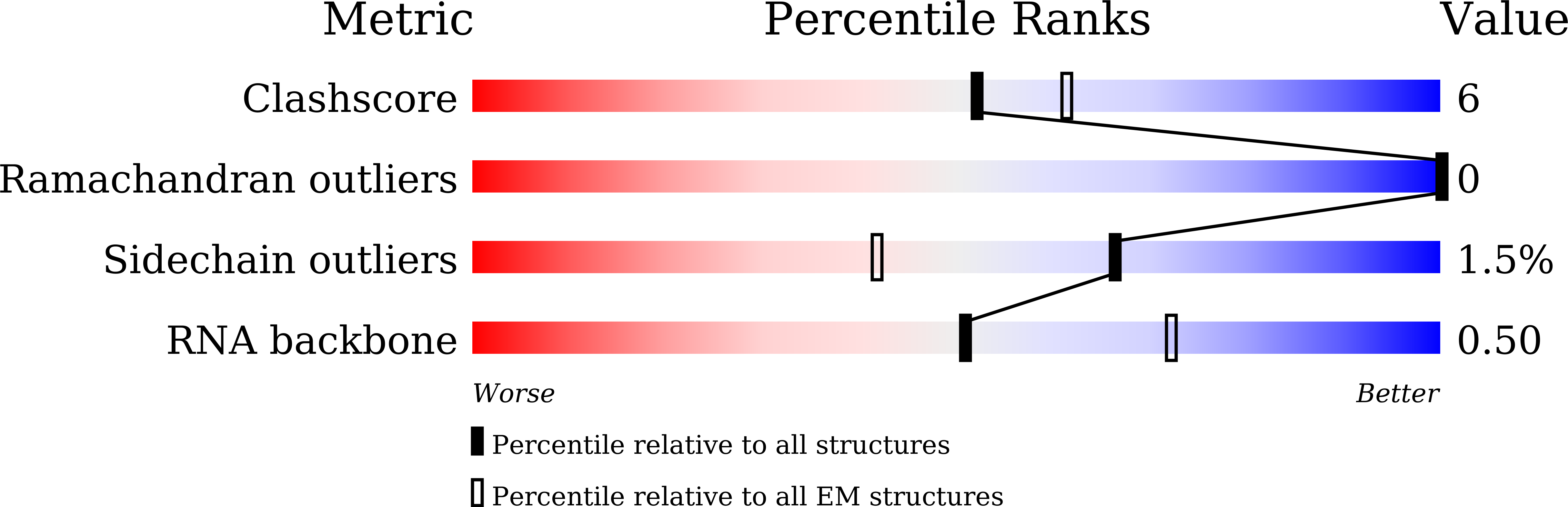
Resolution:
4.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE