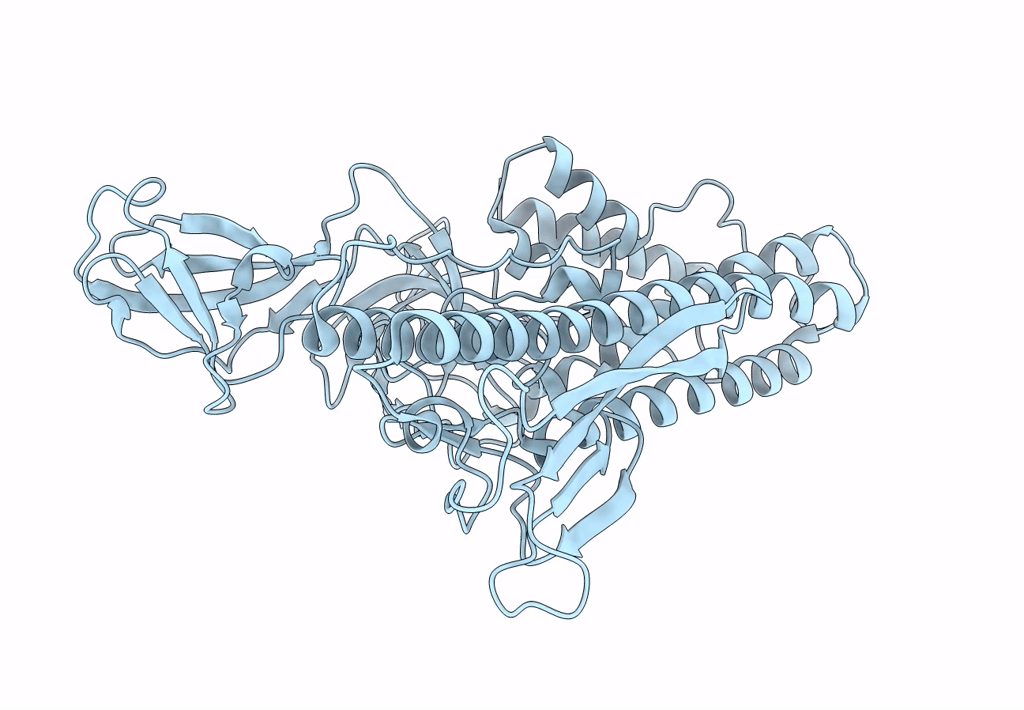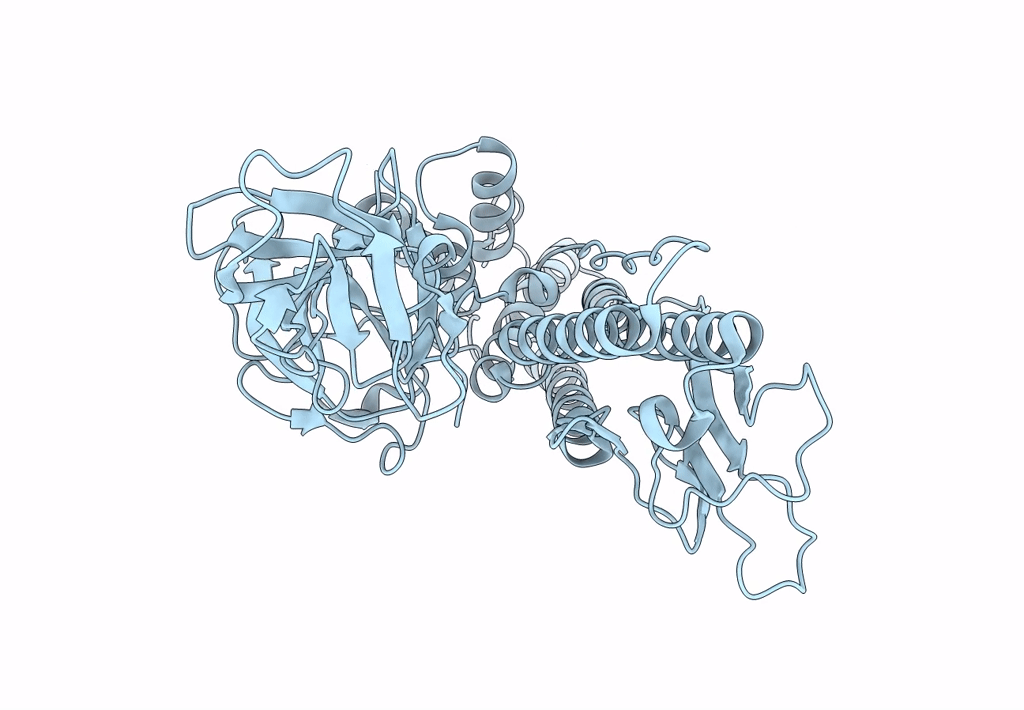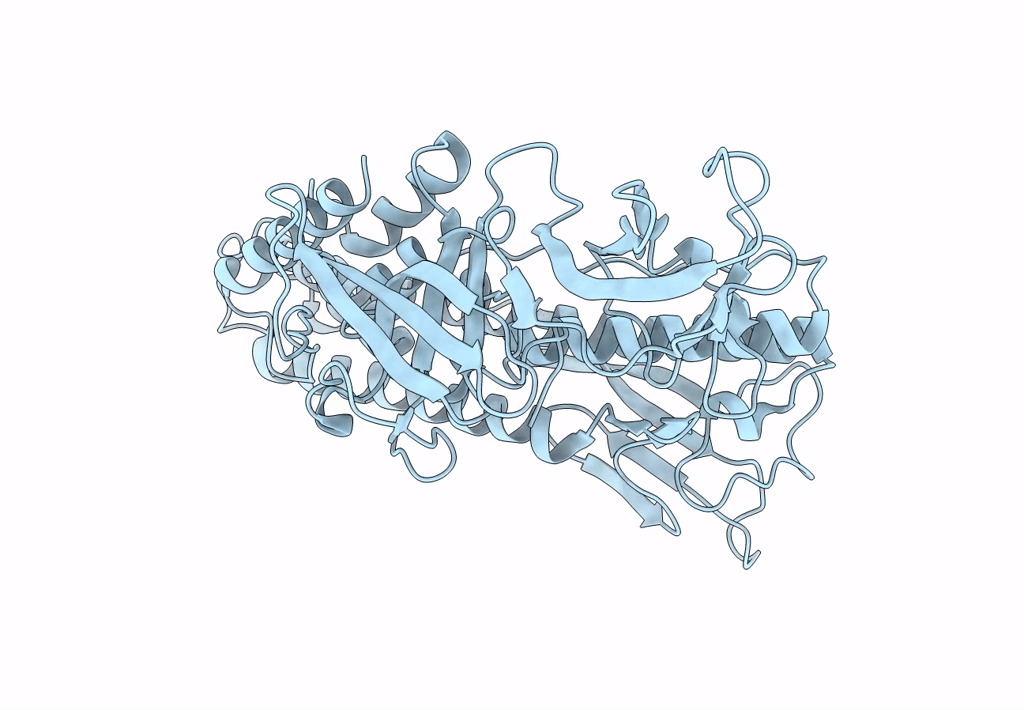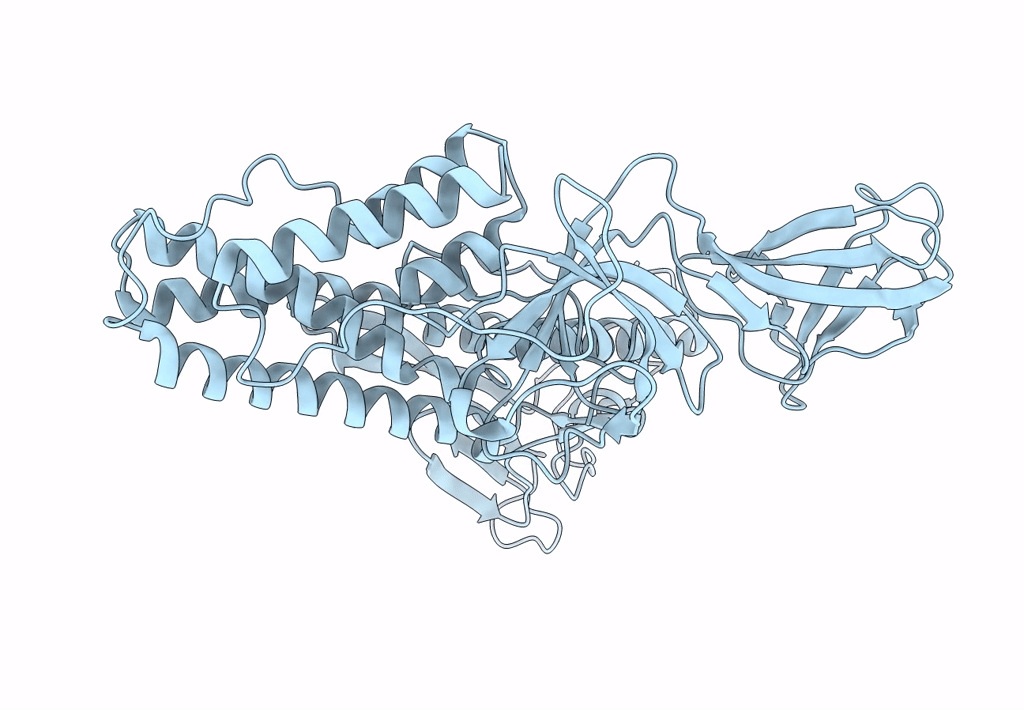
Deposition Date
2019-01-18
Release Date
2019-03-06
Last Version Date
2024-03-27
Entry Detail
PDB ID:
6J7V
Keywords:
Title:
Structure of HRPV6 VP5 fitted in the cryoEM density of the spike
Biological Source:
Source Organism(s):
Halorubrum pleomorphic virus 6 (Taxon ID: 1156721)
Method Details:
Experimental Method:
Resolution:
16.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SUBTOMOGRAM AVERAGING


