Deposition Date
2019-01-08
Release Date
2020-01-15
Last Version Date
2024-03-27
Entry Detail
PDB ID:
6J4B
Keywords:
Title:
Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 400 mM Zinc acetate
Biological Source:
Source Organism(s):
Streptomyces sp. B9173 (Taxon ID: 1462558)
Expression System(s):
Method Details:
Experimental Method:
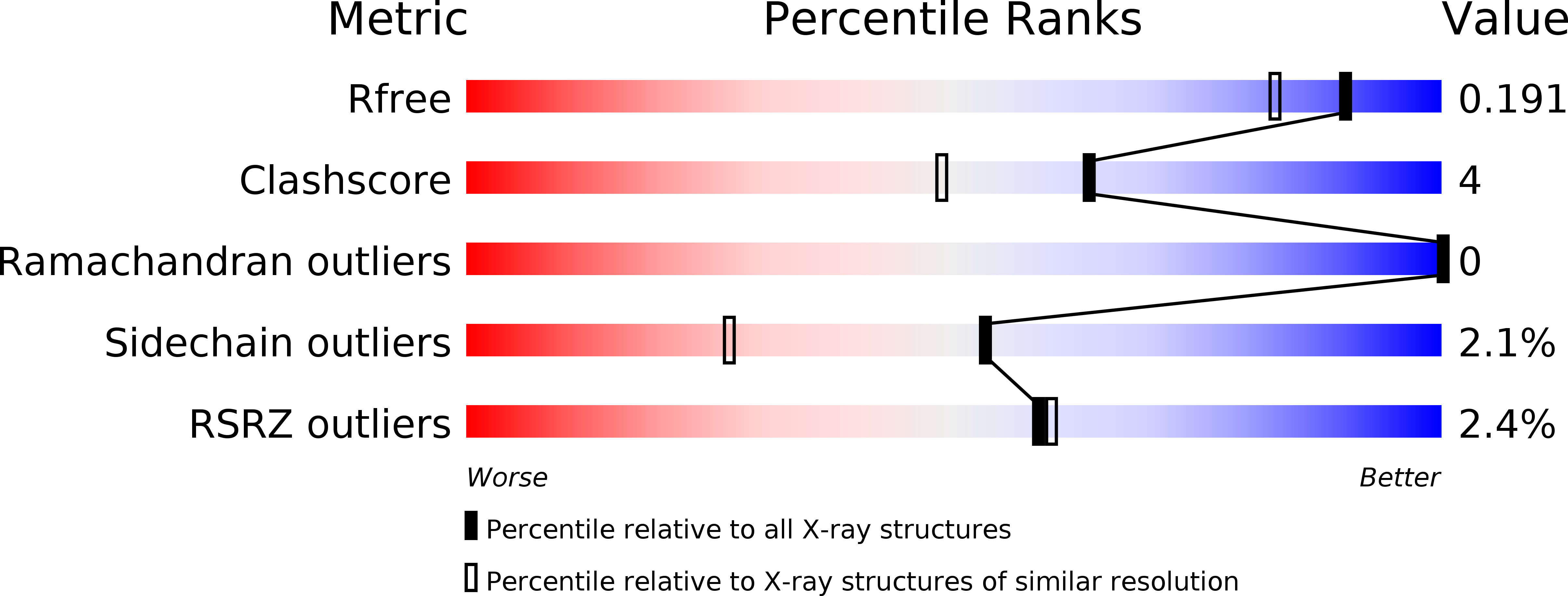
Resolution:
1.58 Å
R-Value Free:
0.17
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
P 32 2 1