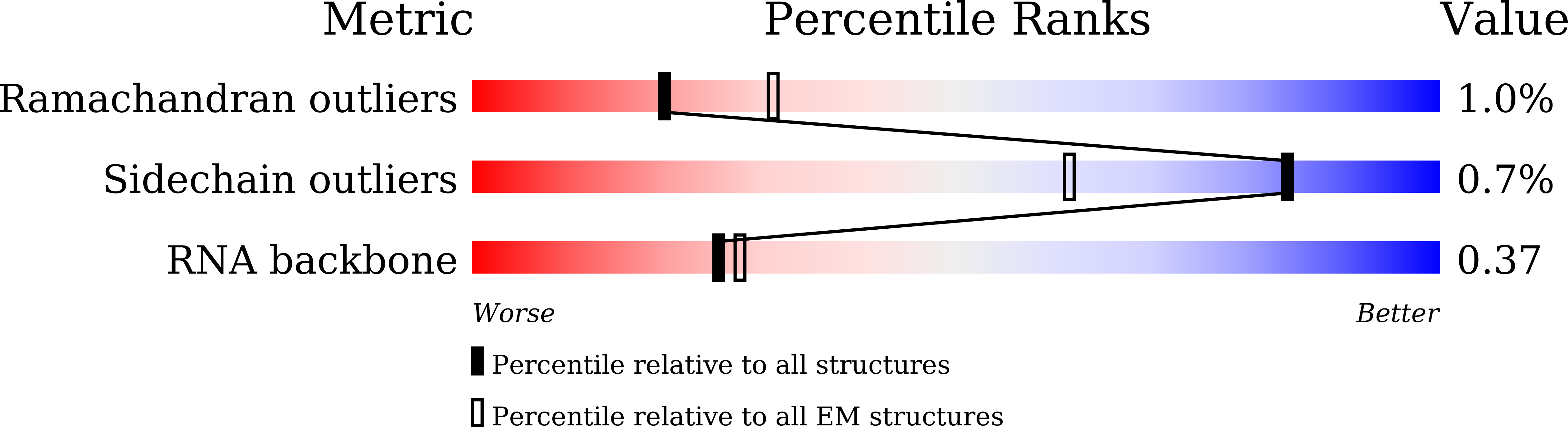
Deposition Date
2018-10-26
Release Date
2019-04-03
Last Version Date
2024-03-27
Entry Detail
PDB ID:
6INQ
Keywords:
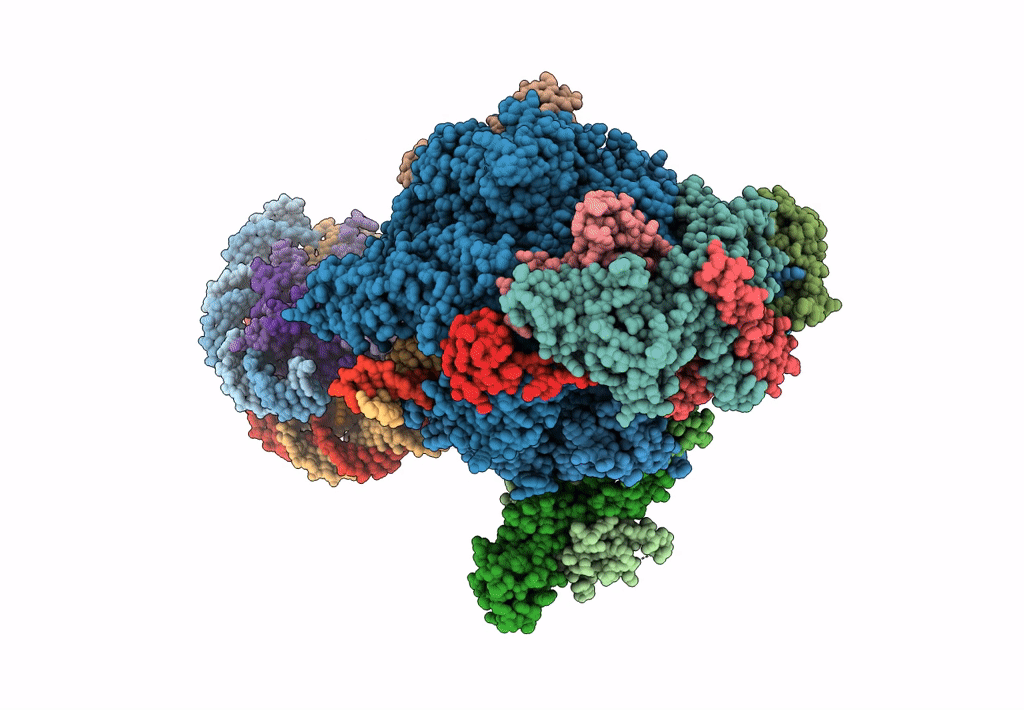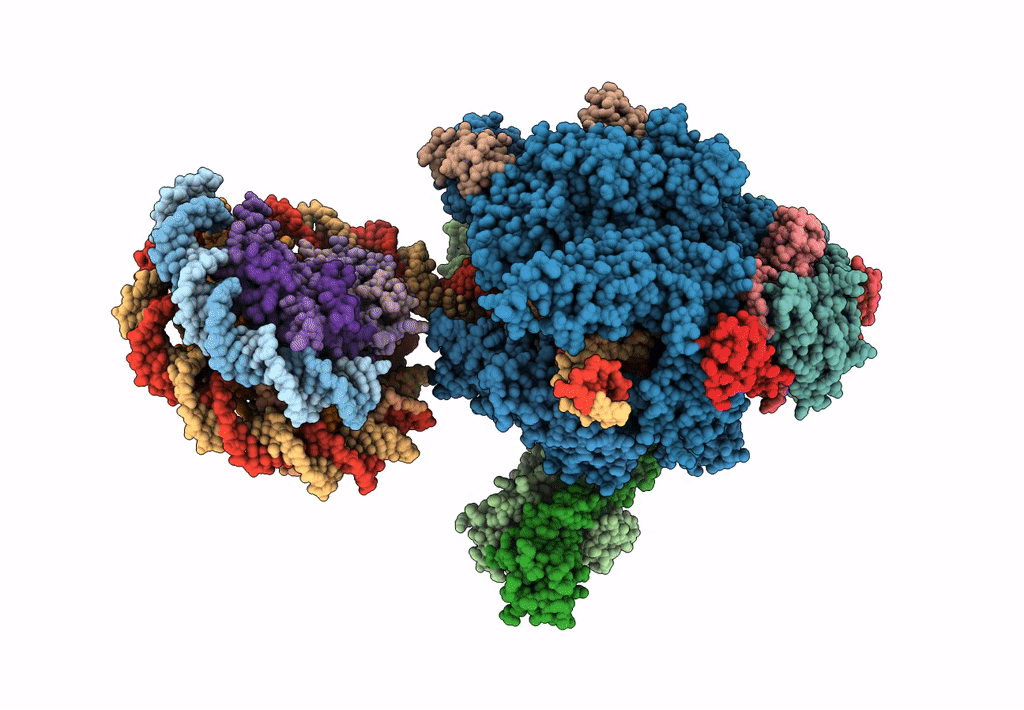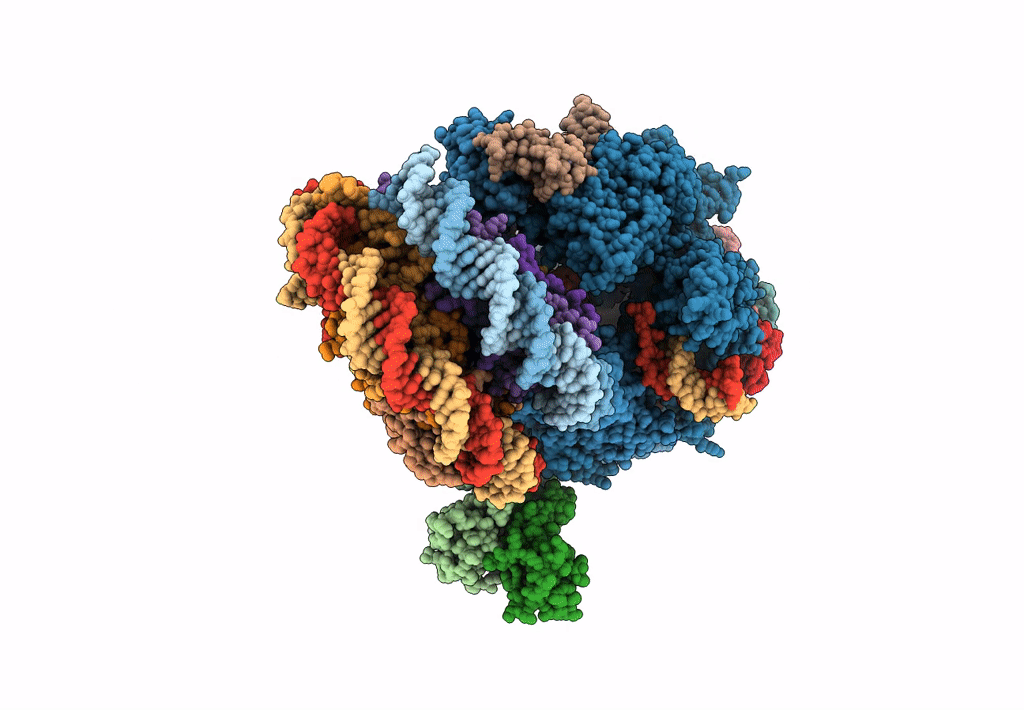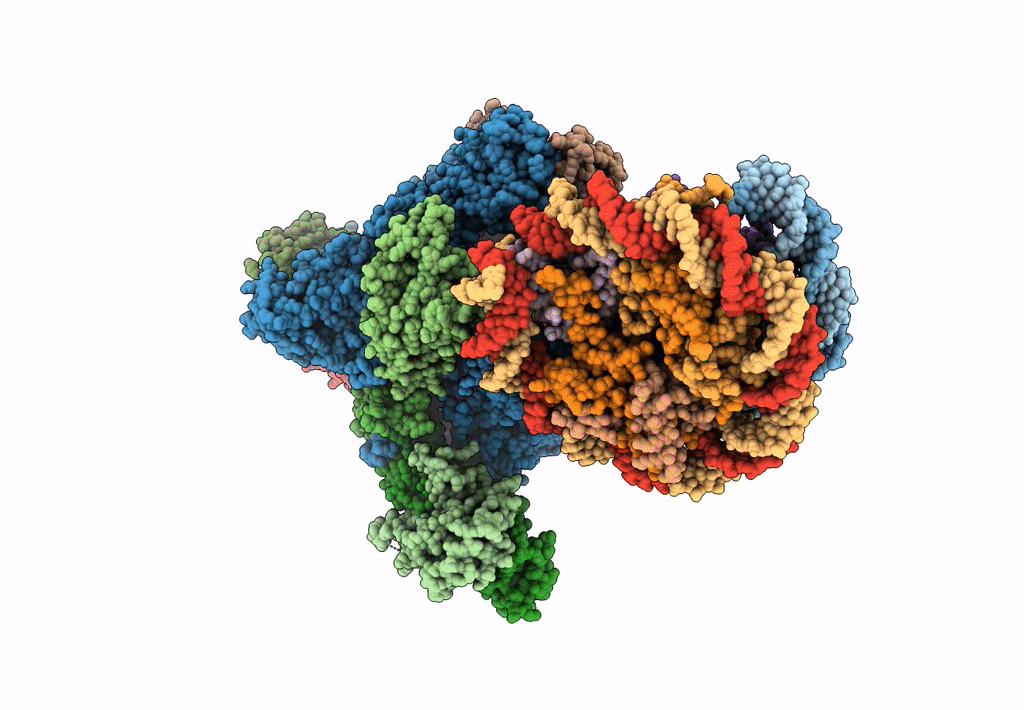
Title:
RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position)
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Komagataella phaffii (strain GS115 / ATCC 20864) (Taxon ID: 644223)
synthetic construct (Taxon ID: 32630)
Komagataella phaffii (strain GS115 / ATCC 20864) (Taxon ID: 644223)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
6.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
CRYSTALLOGRAPHY