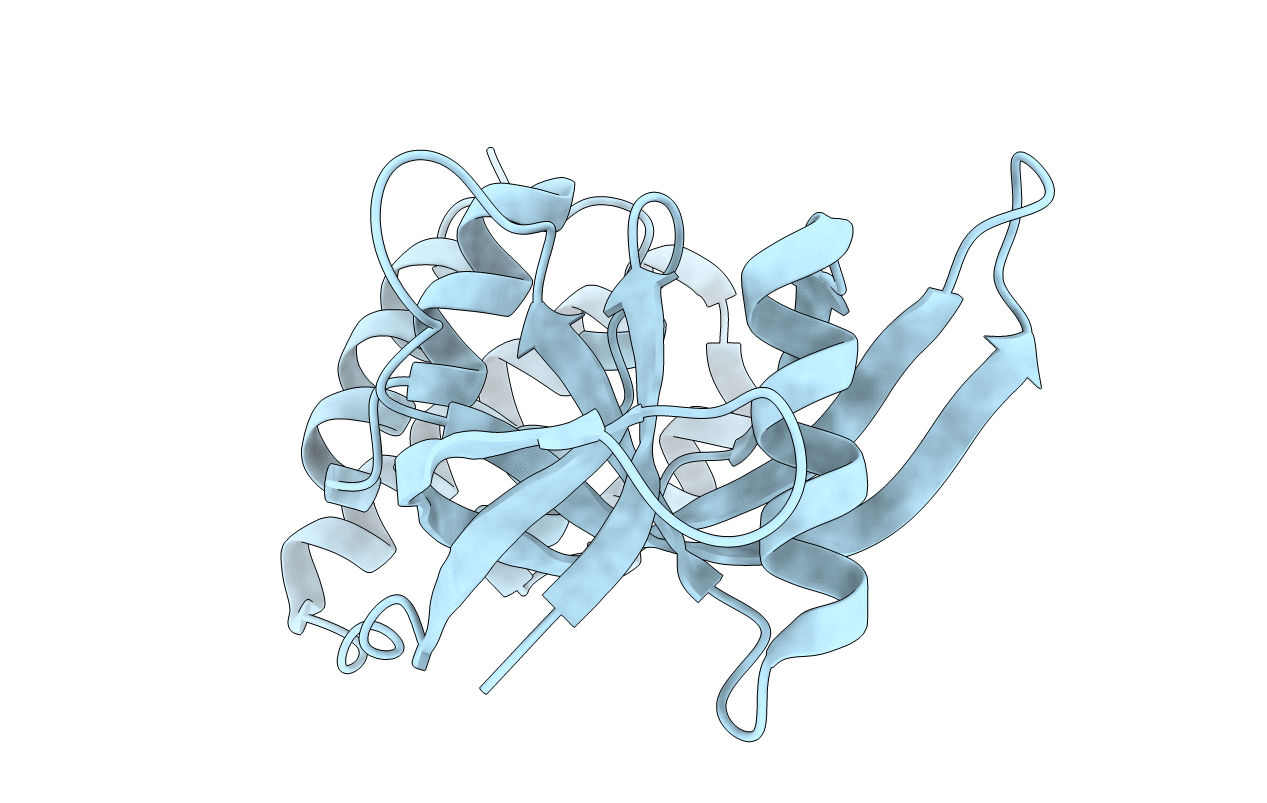
Deposition Date
2018-09-29
Release Date
2019-08-07
Last Version Date
2023-11-22
Entry Detail
PDB ID:
6IHG
Keywords:
Title:
N terminal domain of Mycobacterium avium complex Lon protease
Biological Source:
Source Organism:
Mycobacterium [tuberculosis] TKK-01-0051 (Taxon ID: 1324261)
Host Organism:
Method Details:
Experimental Method:
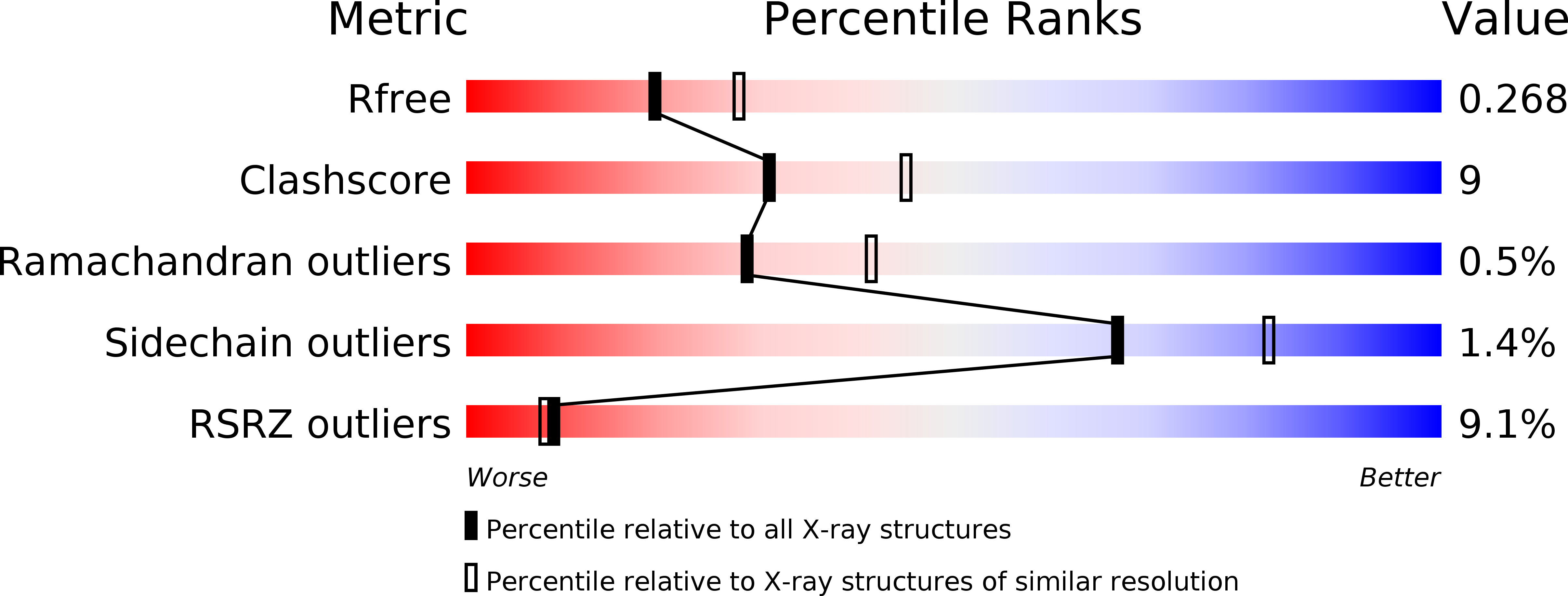
Resolution:
2.40 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 31 2 1