Abstact
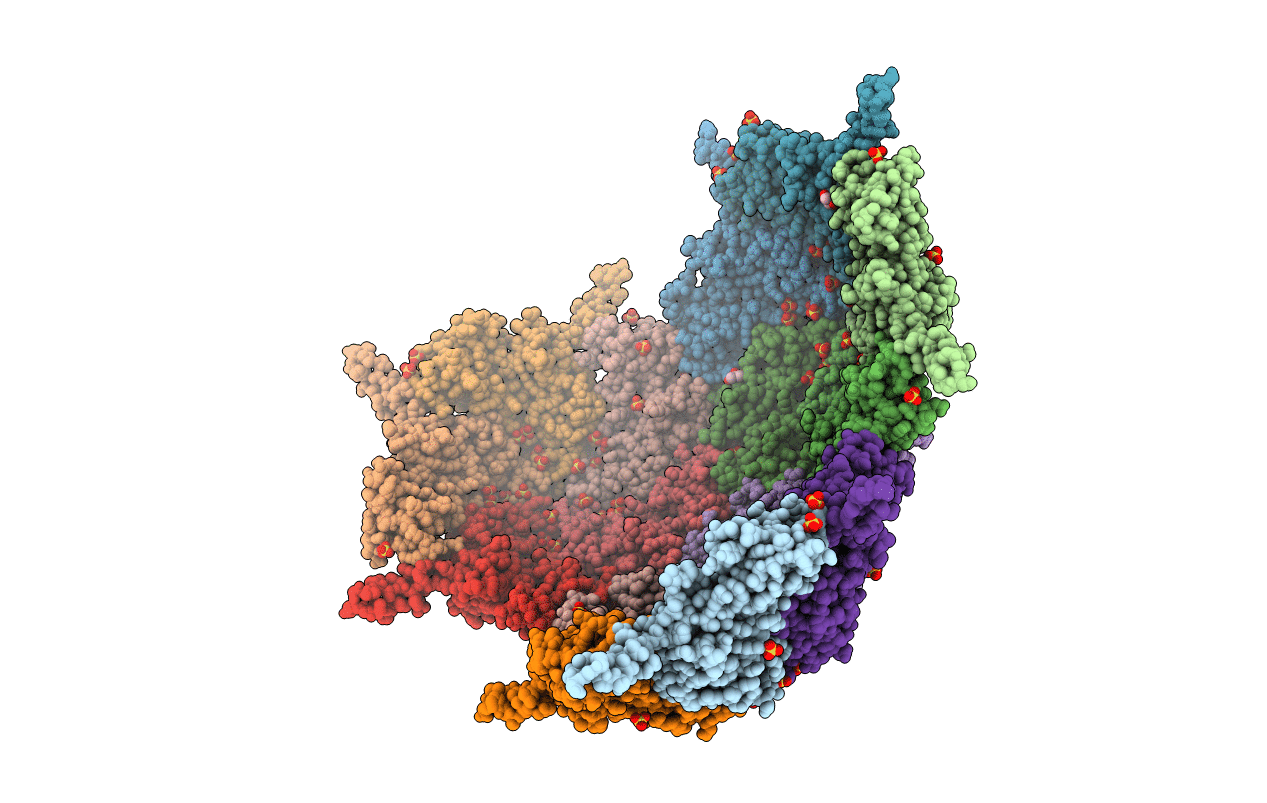
Using a newly discovered encapsulin from Mycolicibacterium hassiacum, several biocatalysts were packaged in this robust protein cage. The encapsulin was found to be easy to produce as recombinant protein. Elucidation of its crystal structure revealed that it is a spherical protein cage of 60 protomers (diameter of 23 nm) with narrow pores. By developing an effective coexpression and isolation procedure, the effect of packaging a variety of biocatalysts could be evaluated. It was shown that encapsulation results in a significantly higher stability of the biocatalysts. Most of the targeted cofactor-containing biocatalysts remained active in the encapsulin. Due to the restricted diameters of the encapsulin pores (5-9 Å), the protein cage protects the encapsulated enzymes from bulky compounds. The work shows that encapsulins may be valuable tools to tune the properties of biocatalysts such as stability and substrate specificity.