Deposition Date
2018-11-19
Release Date
2019-01-30
Last Version Date
2024-01-24
Entry Detail
PDB ID:
6I8A
Keywords:
Title:
The crystal structure of the Pol2 catalytic domain of DNA polymerase epsilon carrying a P301R substitution.
Biological Source:
Source Organism:
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Taxon ID: 559292)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
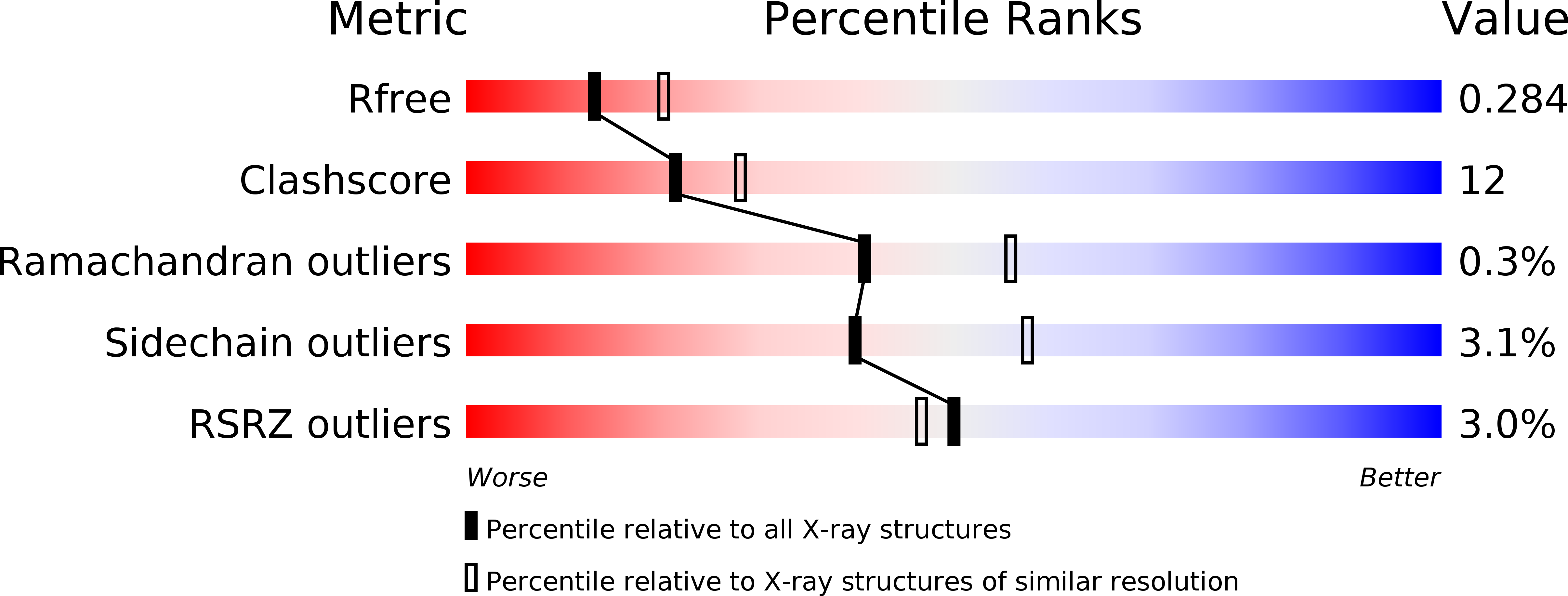
Resolution:
2.65 Å
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.23
Space Group:
P 1 2 1