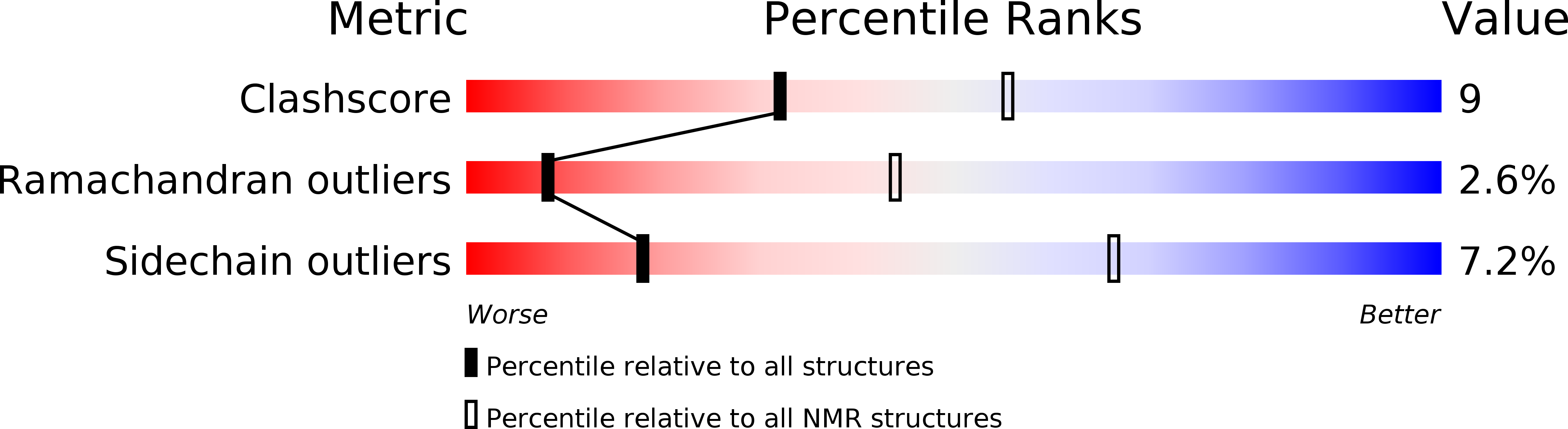Deposition Date
2018-11-07
Release Date
2019-02-20
Last Version Date
2024-06-19
Entry Detail
PDB ID:
6I3R
Keywords:
Title:
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1/2 of Drosophila helicase MLE
Biological Source:
Source Organism(s):
Drosophila melanogaster (Taxon ID: 7227)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
10
Conformers Submitted:
10
Selection Criteria:
structures with the lowest energy