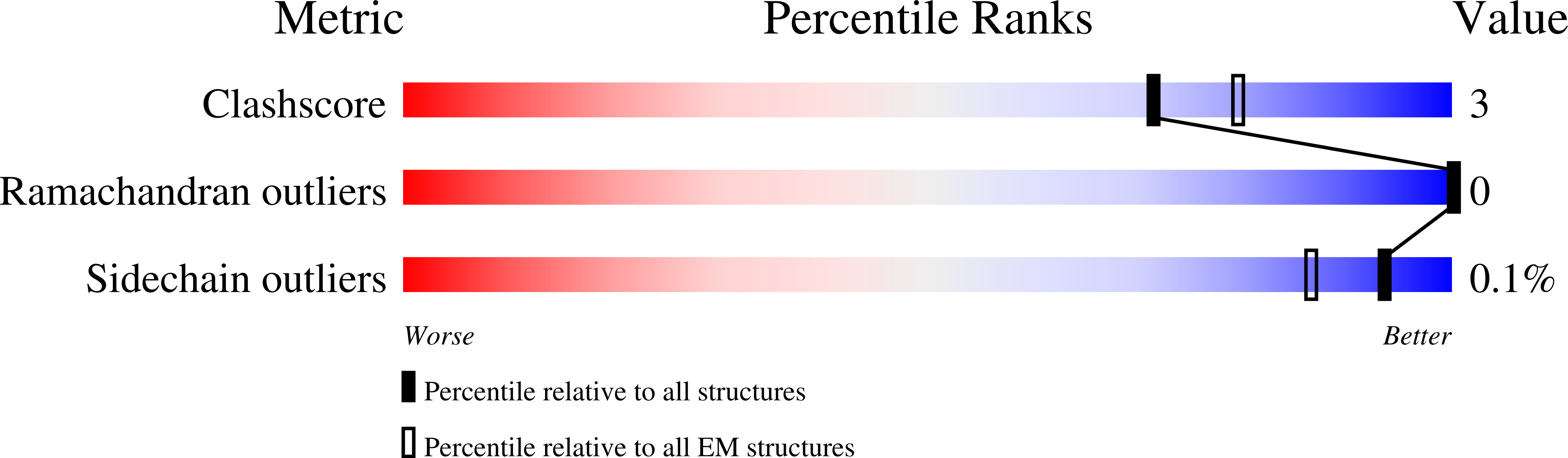
Deposition Date
2018-10-18
Release Date
2019-06-12
Last Version Date
2025-07-09
Entry Detail
PDB ID:
6HY0
Keywords:
Title:
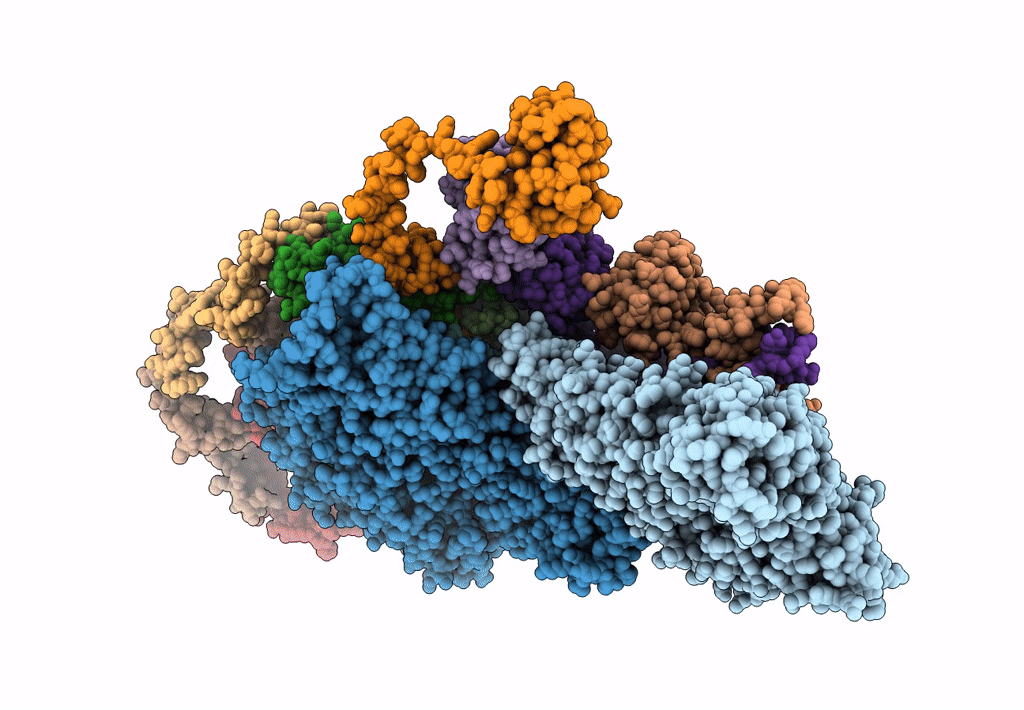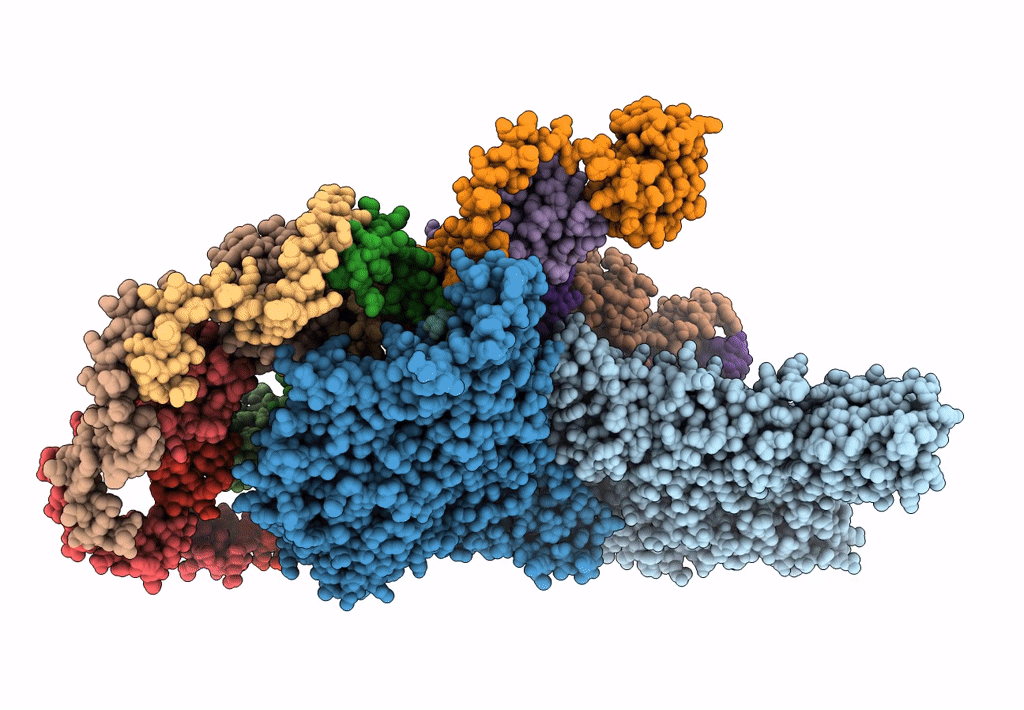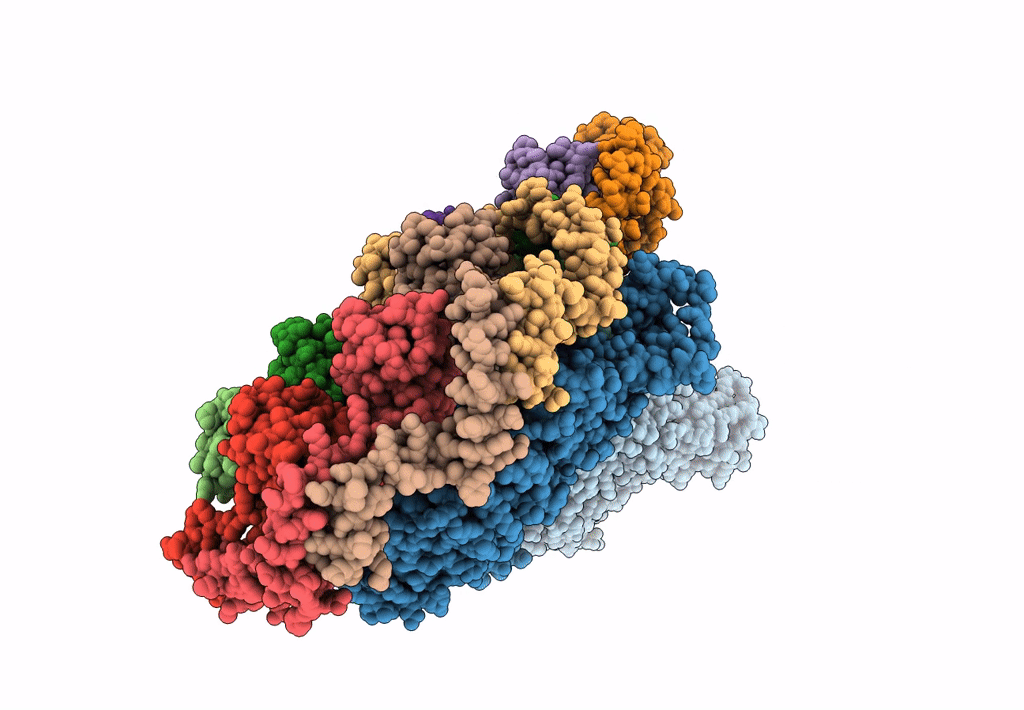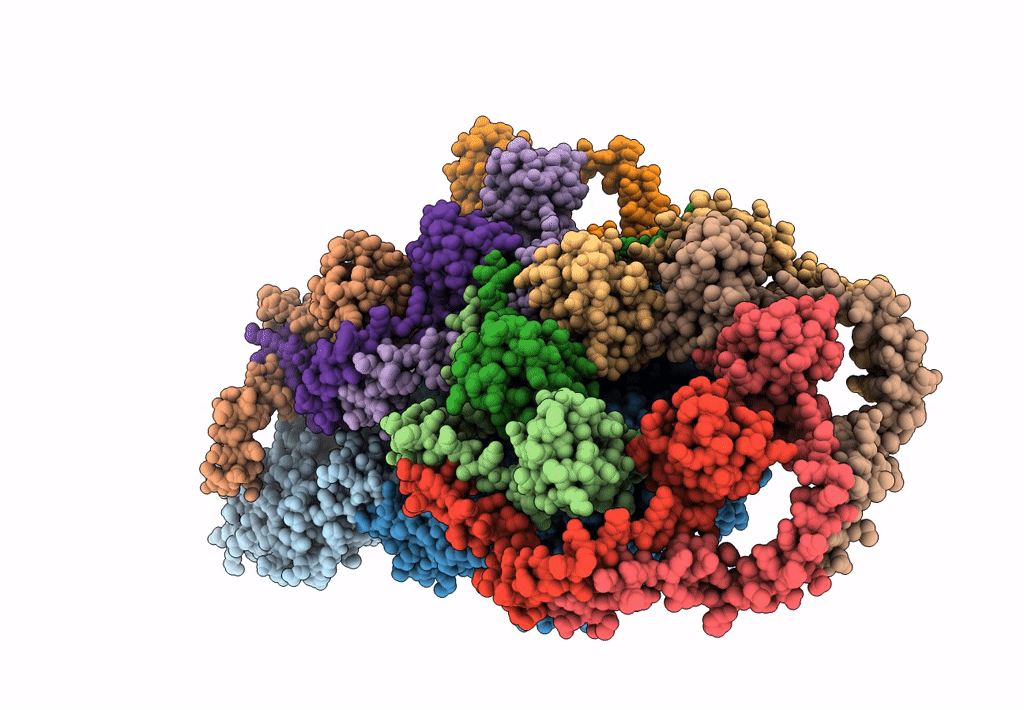
Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry
Biological Source:
Source Organism:
Pseudomonas phage phi6 (Taxon ID: 10879)
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE