Deposition Date
2018-09-11
Release Date
2019-03-06
Last Version Date
2024-01-24
Entry Detail
PDB ID:
6HM1
Keywords:
Title:
Structural and thermodynamic signatures of ligand binding to an enigmatic chitinase-D from Serratia proteamaculans
Biological Source:
Source Organism(s):
Serratia proteamaculans (strain 568) (Taxon ID: 399741)
Expression System(s):
Method Details:
Experimental Method:
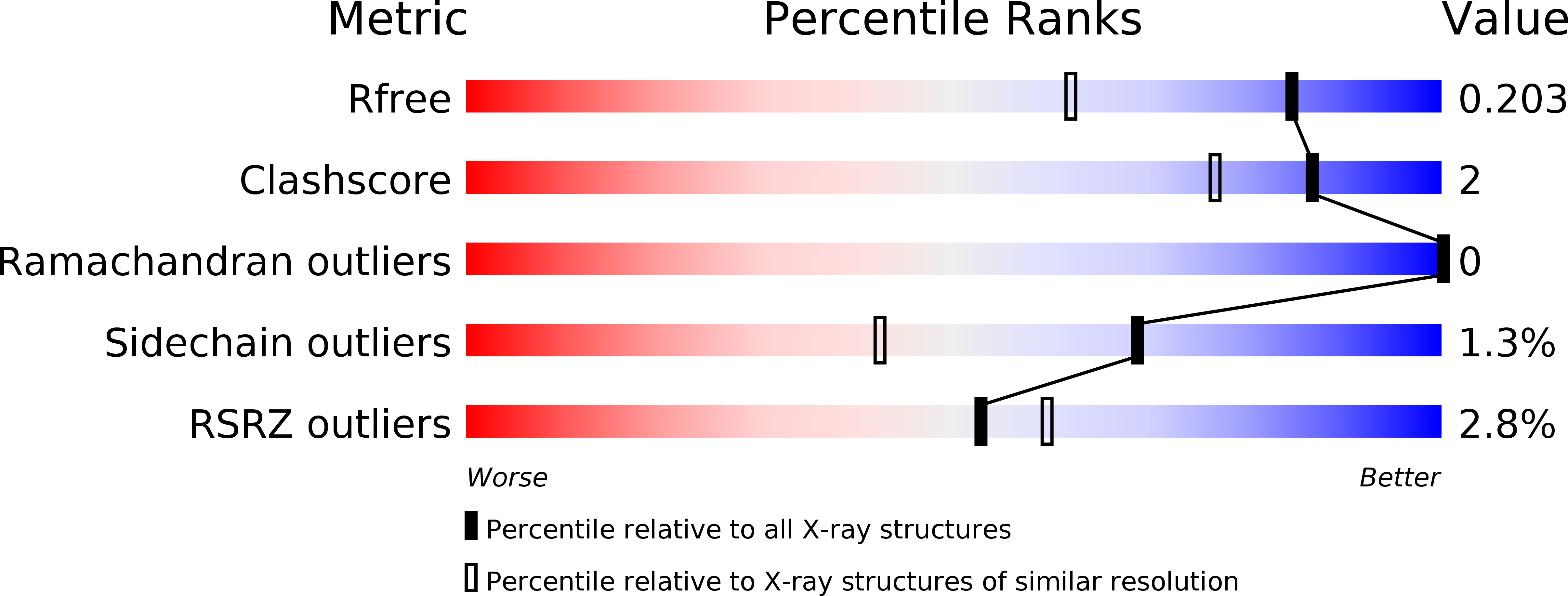
Resolution:
1.54 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.17
Space Group:
P 21 21 21