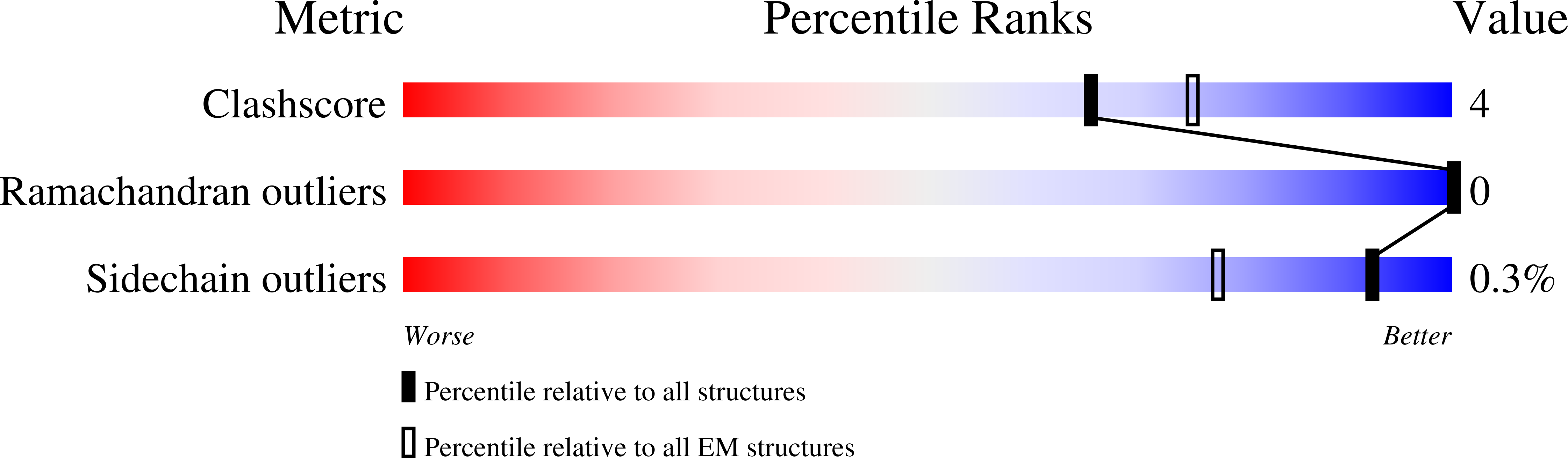
Deposition Date
2018-08-30
Release Date
2018-11-07
Last Version Date
2024-11-20
Entry Detail
PDB ID:
6HIQ
Keywords:
Title:
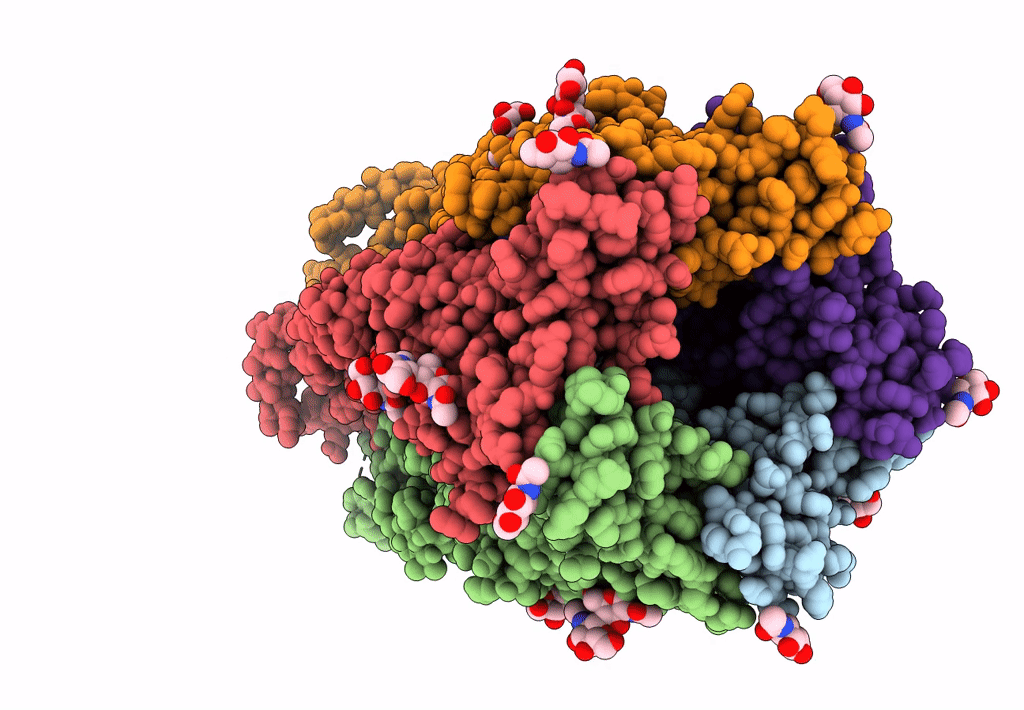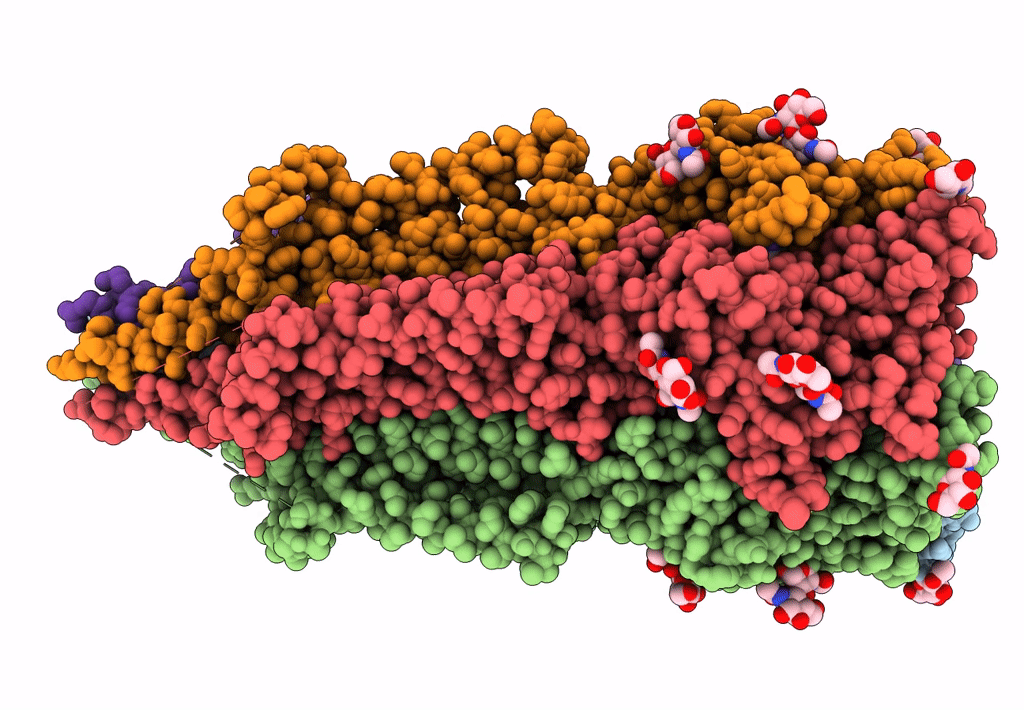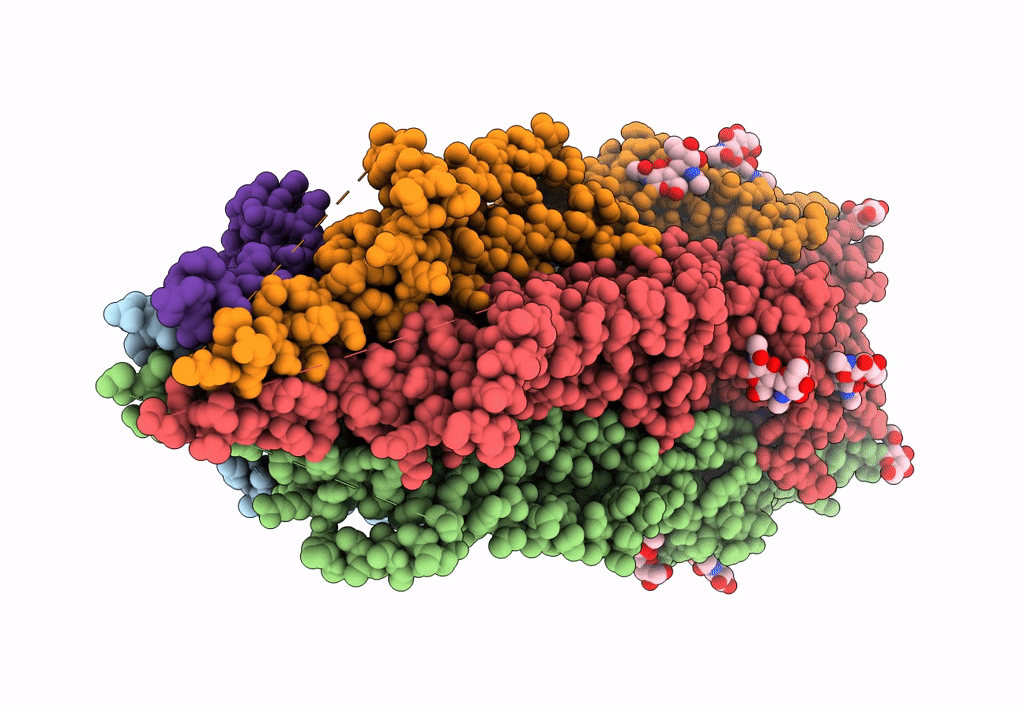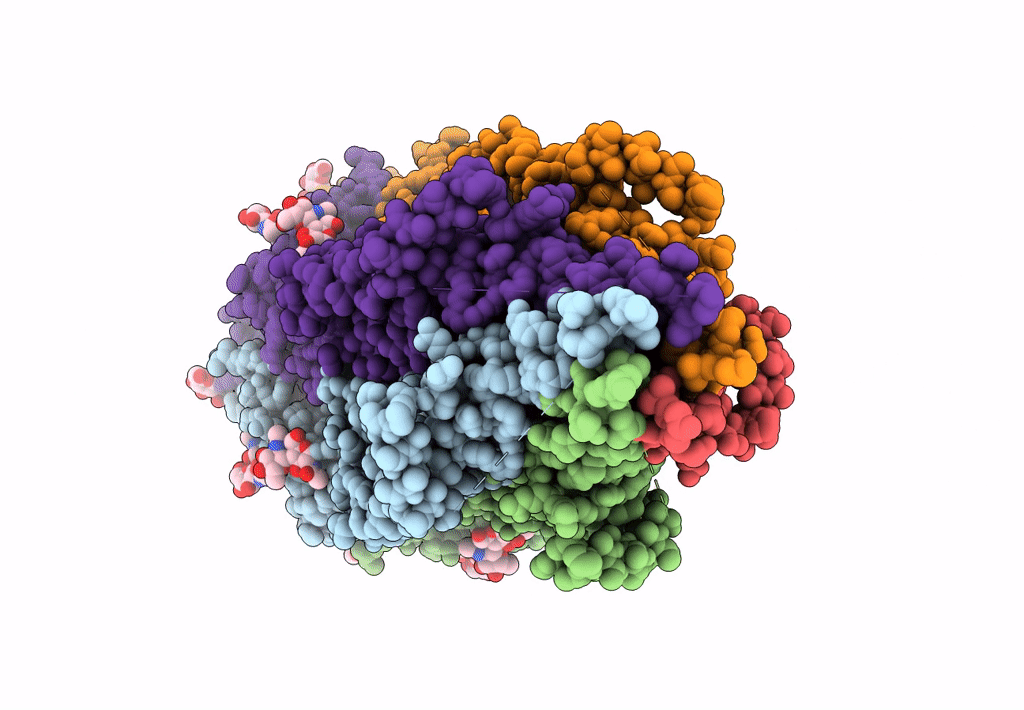
Mouse serotonin 5-HT3 receptor, serotonin-bound, I2 conformation
Biological Source:
Source Organism:
Mus musculus (Taxon ID: 10090)
Host Organism:
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE