Deposition Date
2018-08-10
Release Date
2018-12-12
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6HBA
Keywords:
Title:
Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form
Biological Source:
Source Organism:
Synechococcus elongatus (strain PCC 7942) (Taxon ID: 1140)
Host Organism:
Method Details:
Experimental Method:
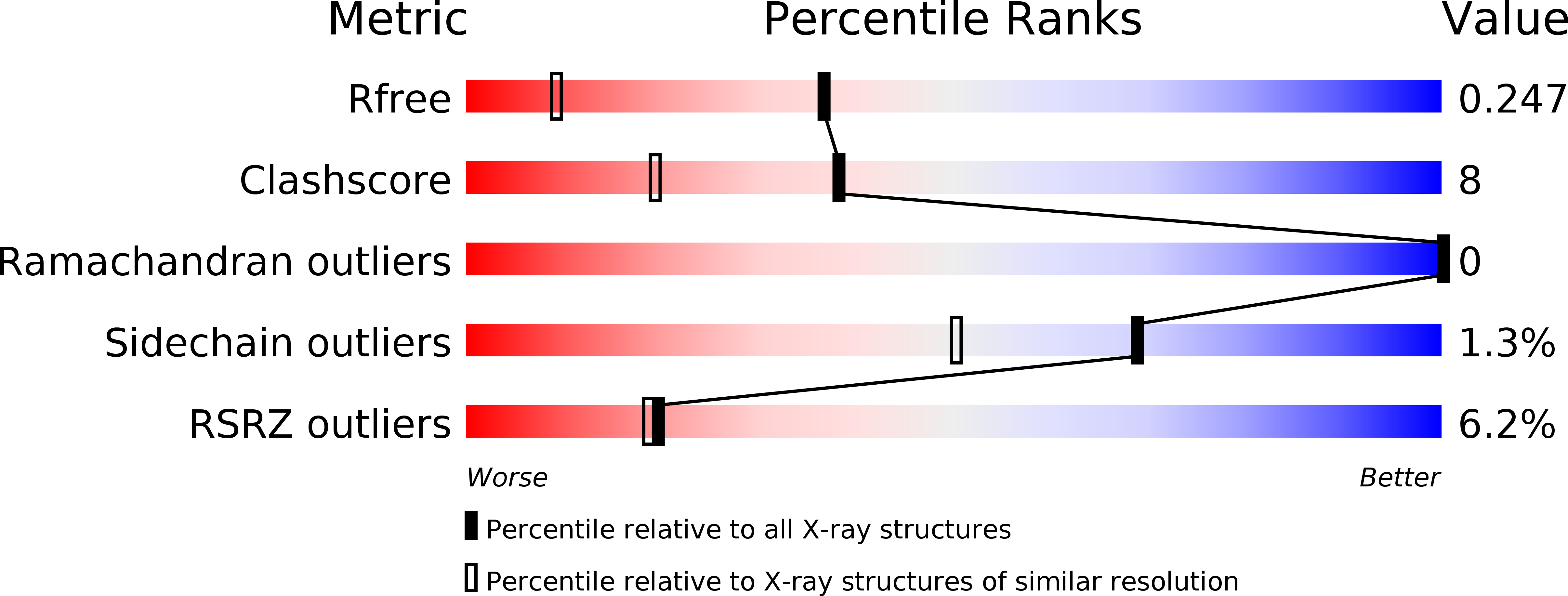
Resolution:
1.65 Å
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 1 21 1