Deposition Date
2018-07-25
Release Date
2019-06-12
Last Version Date
2024-05-15
Entry Detail
PDB ID:
6H5S
Keywords:
Title:
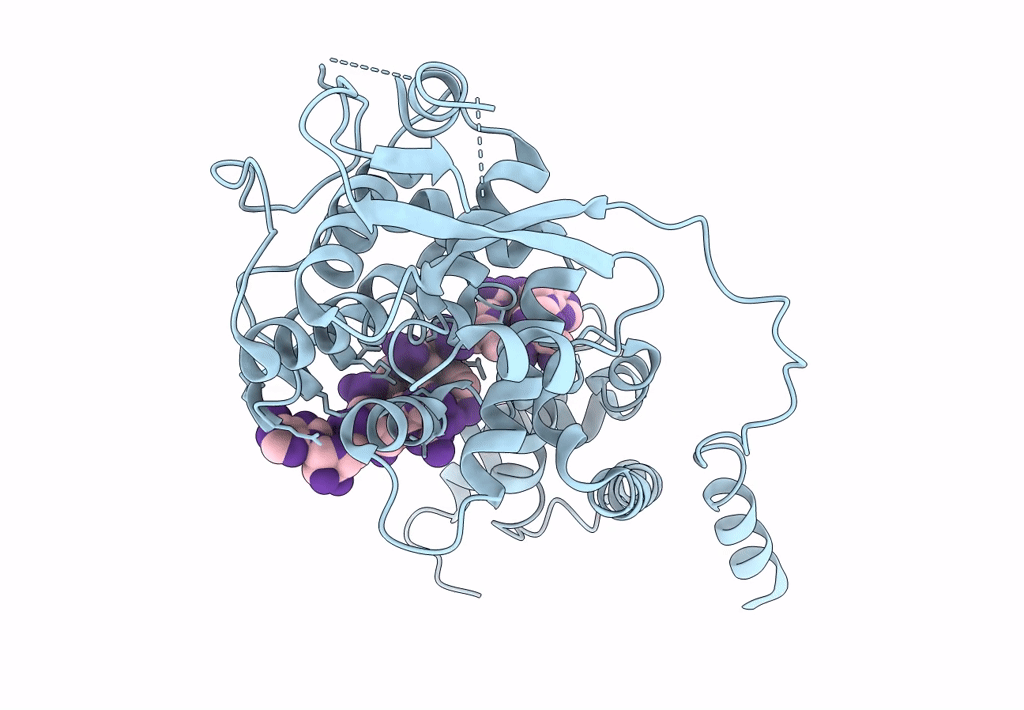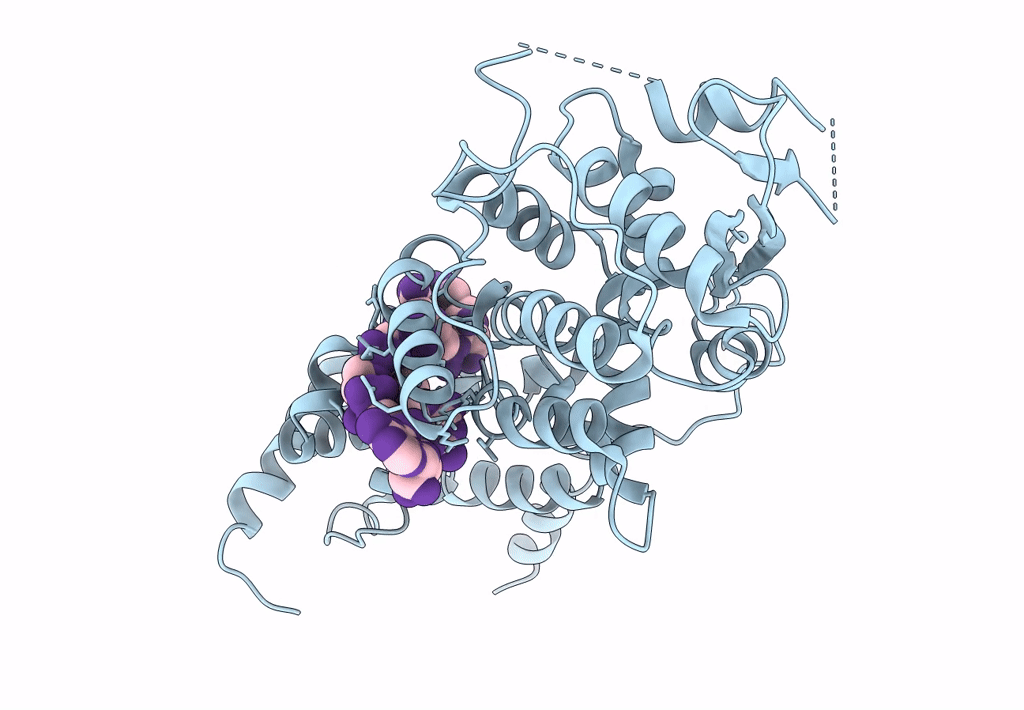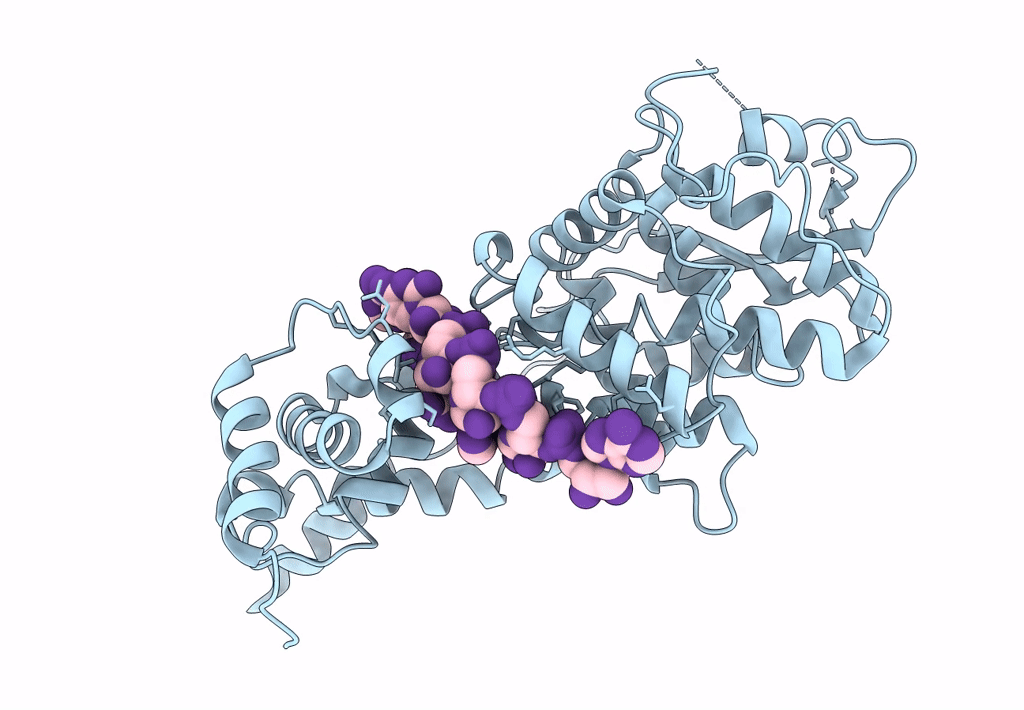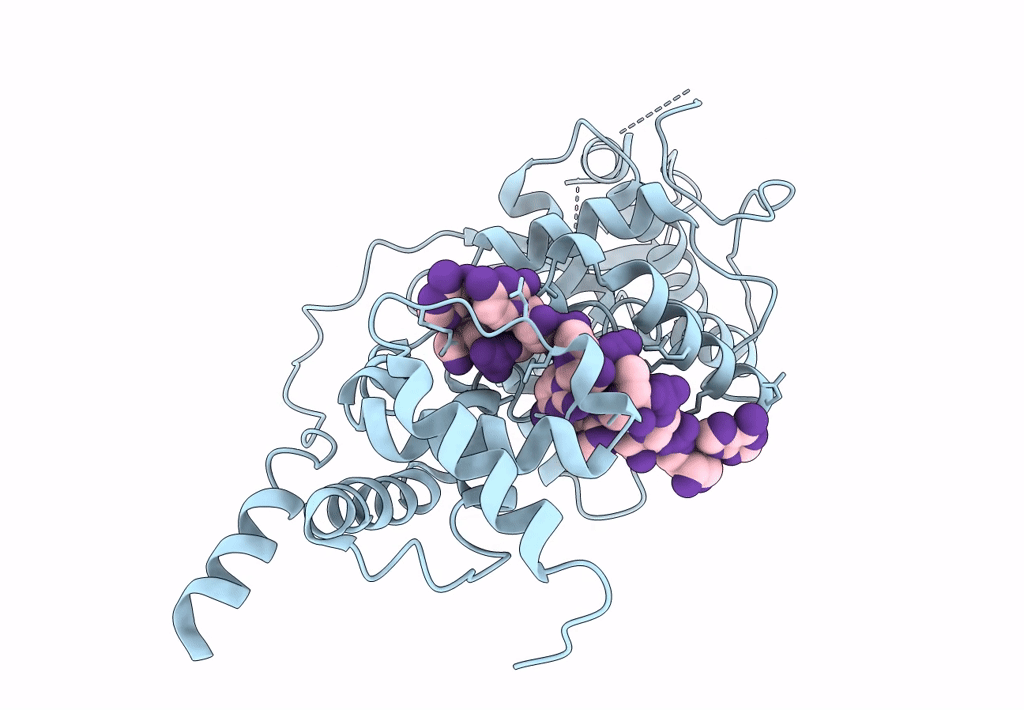
Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers.
Biological Source:
Source Organism(s):
Measles morbillivirus (Taxon ID: 11234)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.30 Å
Aggregation State:
HELICAL ARRAY
Reconstruction Method:
HELICAL


