Deposition Date
2018-05-26
Release Date
2018-08-22
Last Version Date
2024-11-20
Entry Detail
PDB ID:
6GMH
Keywords:
Title:
Structure of activated transcription complex Pol II-DSIF-PAF-SPT6
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Sus scrofa (Taxon ID: 9823)
synthetic construct (Taxon ID: 32630)
Sus scrofa (Taxon ID: 9823)
Host Organism:
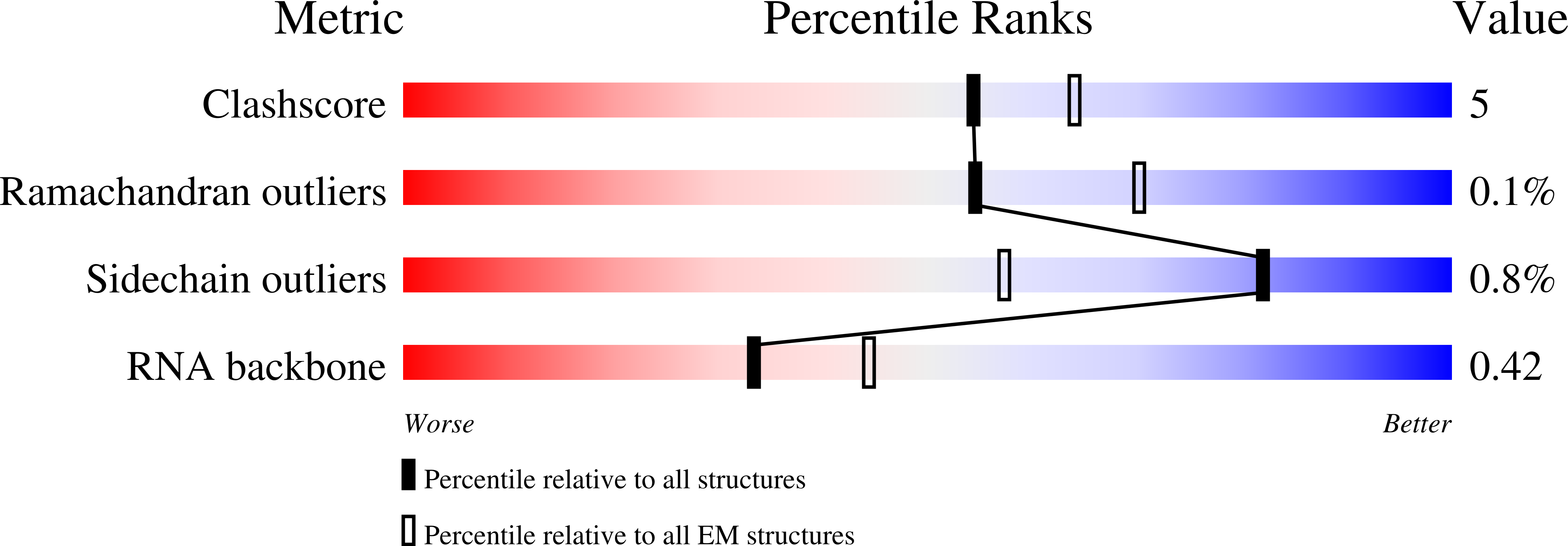
Method Details:
Experimental Method:
Resolution:
3.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE