Deposition Date
2017-12-21
Release Date
2018-01-17
Last Version Date
2024-01-17
Entry Detail
PDB ID:
6FD2
Keywords:
Title:
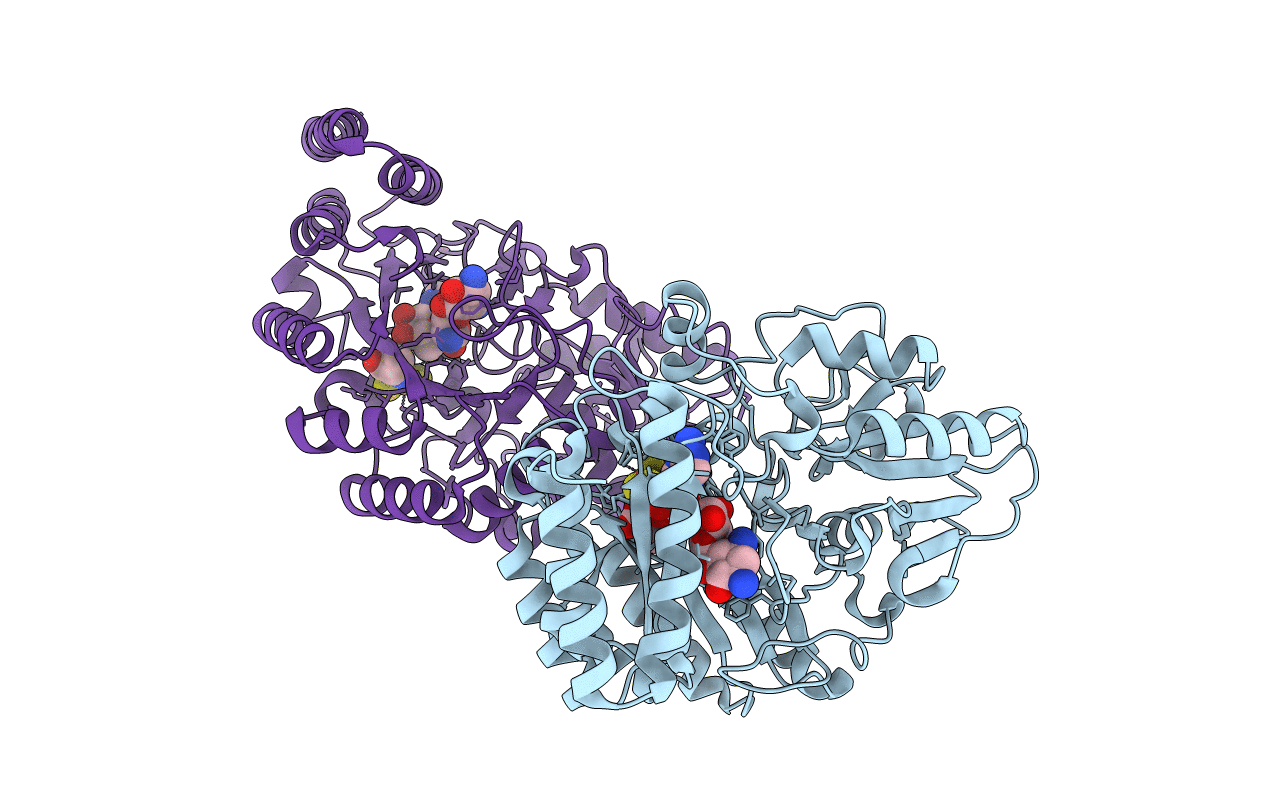
Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
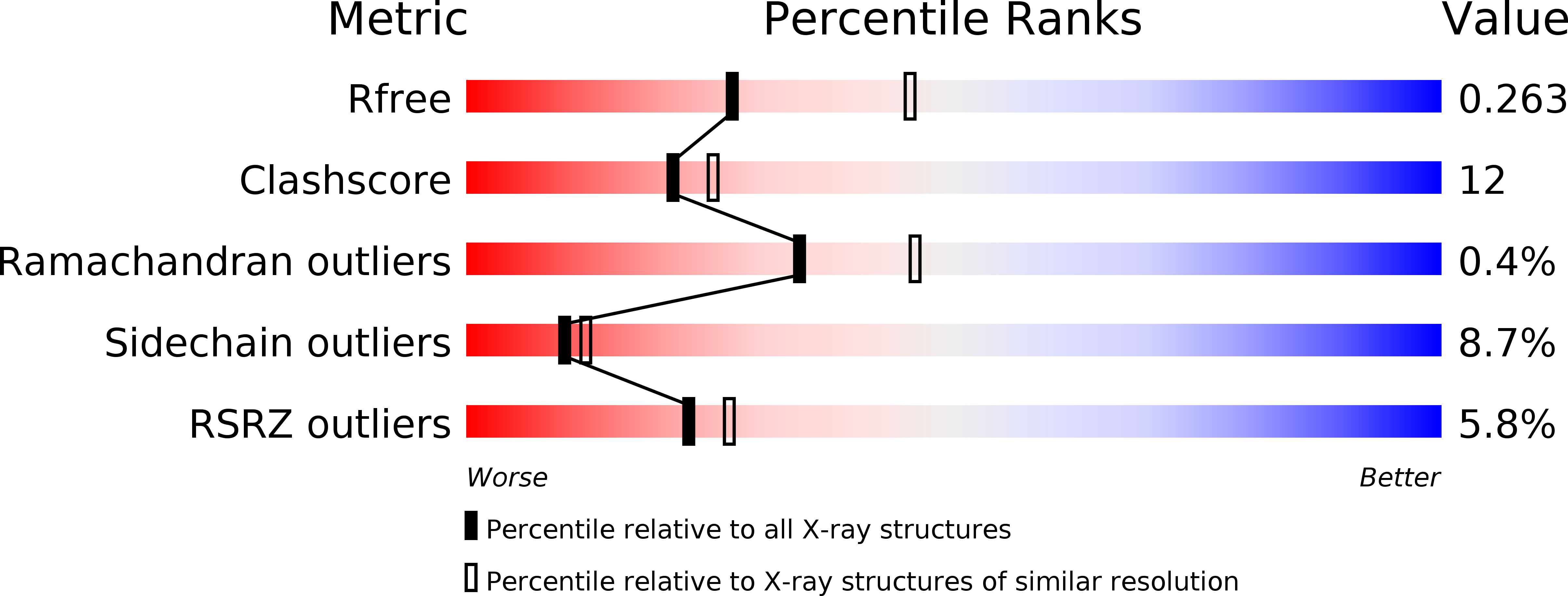
Resolution:
2.55 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 31 2 1