Deposition Date
2017-12-18
Release Date
2018-07-18
Last Version Date
2024-01-17
Entry Detail
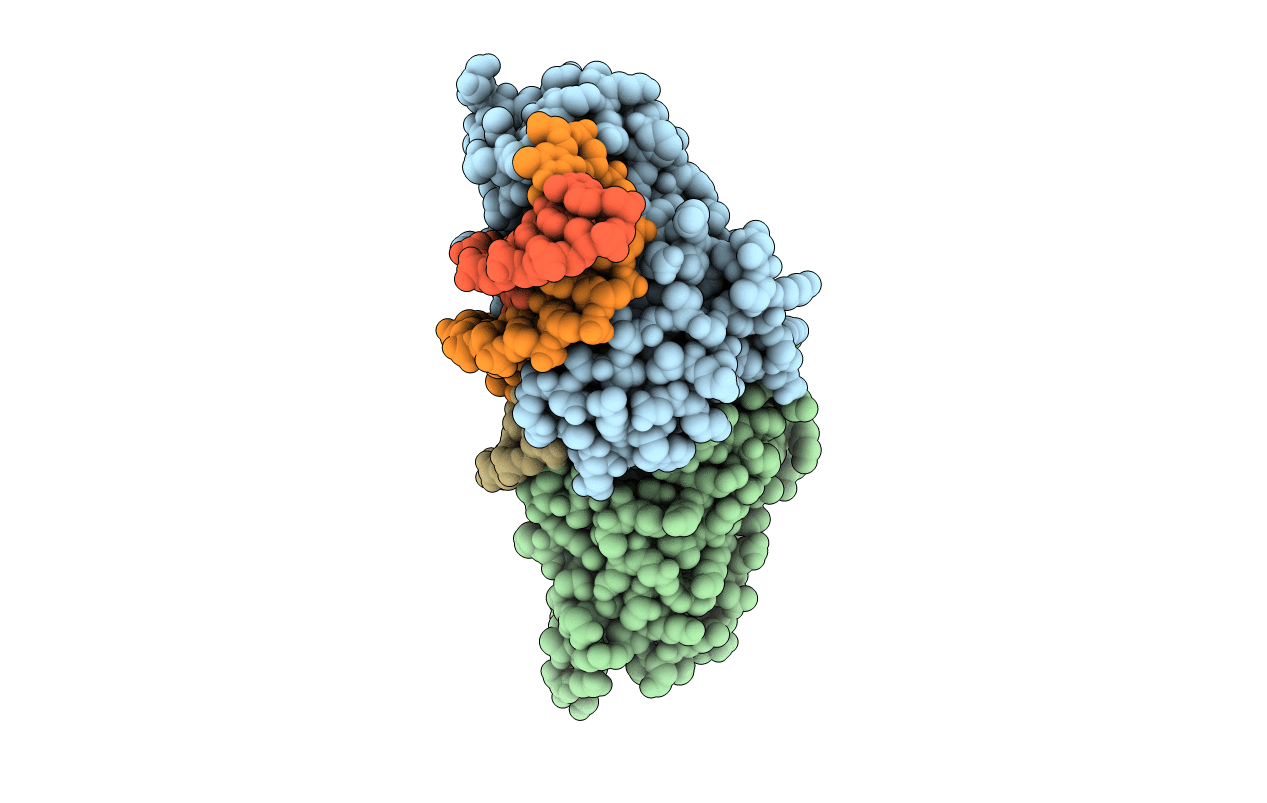
PDB ID:
6FB2
Keywords:
Title:
Crystal Structure of a Tailored I-CreI Homing Endonuclease Protein (3115 variant) in complex with its target DNA (Haemoglobin beta subunit gene) in the presence of Manganese
Biological Source:
Source Organism(s):
Chlamydomonas reinhardtii (Taxon ID: 3055)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.95 Å
R-Value Free:
0.23
R-Value Work:
0.18
R-Value Observed:
0.19
Space Group:
P 1 21 1


