Deposition Date
2018-06-17
Release Date
2018-09-12
Last Version Date
2024-03-13
Entry Detail
PDB ID:
6DTM
Keywords:
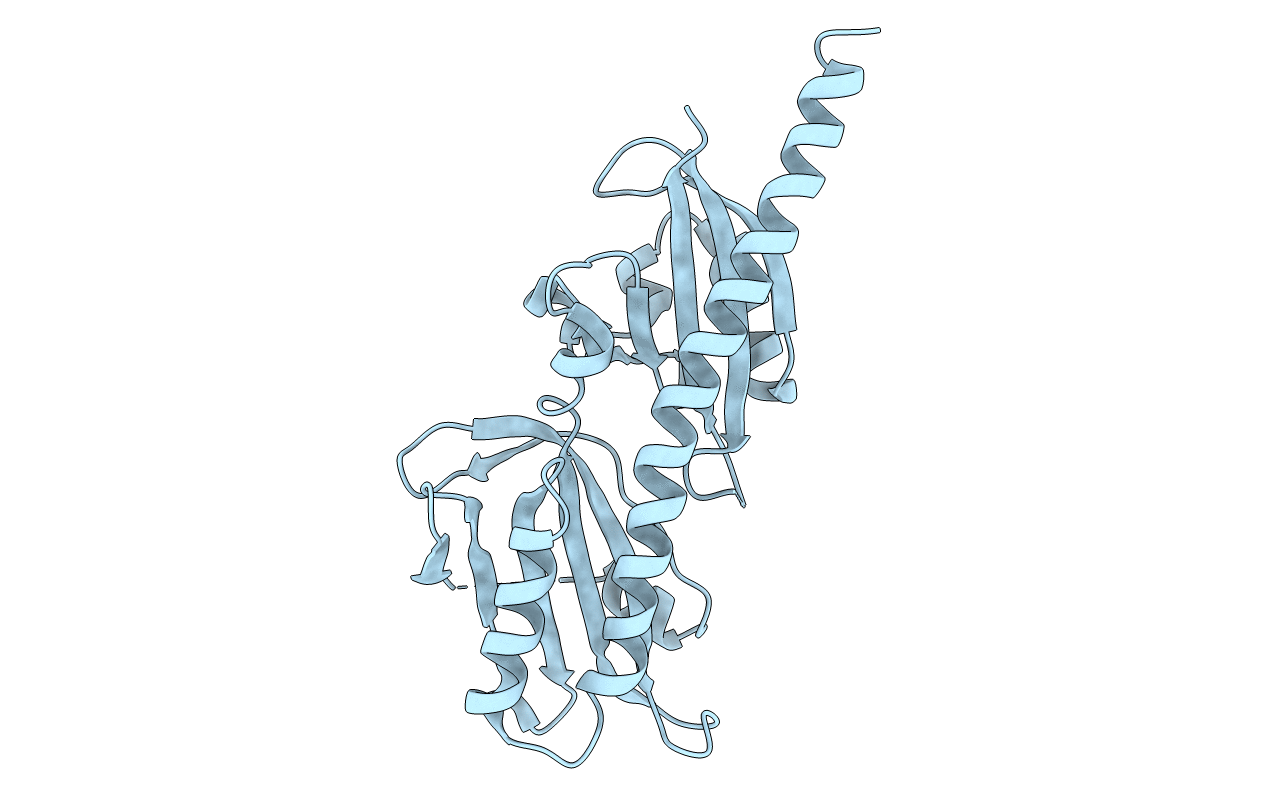
Title:
Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain
Biological Source:
Source Organism(s):
Helicobacter pylori SS1 (Taxon ID: 102617)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.50 Å
R-Value Free:
0.32
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
C 1 2 1


