Deposition Date
2018-02-23
Release Date
2018-03-07
Last Version Date
2024-10-16
Entry Detail
PDB ID:
6CI6
Keywords:
Title:
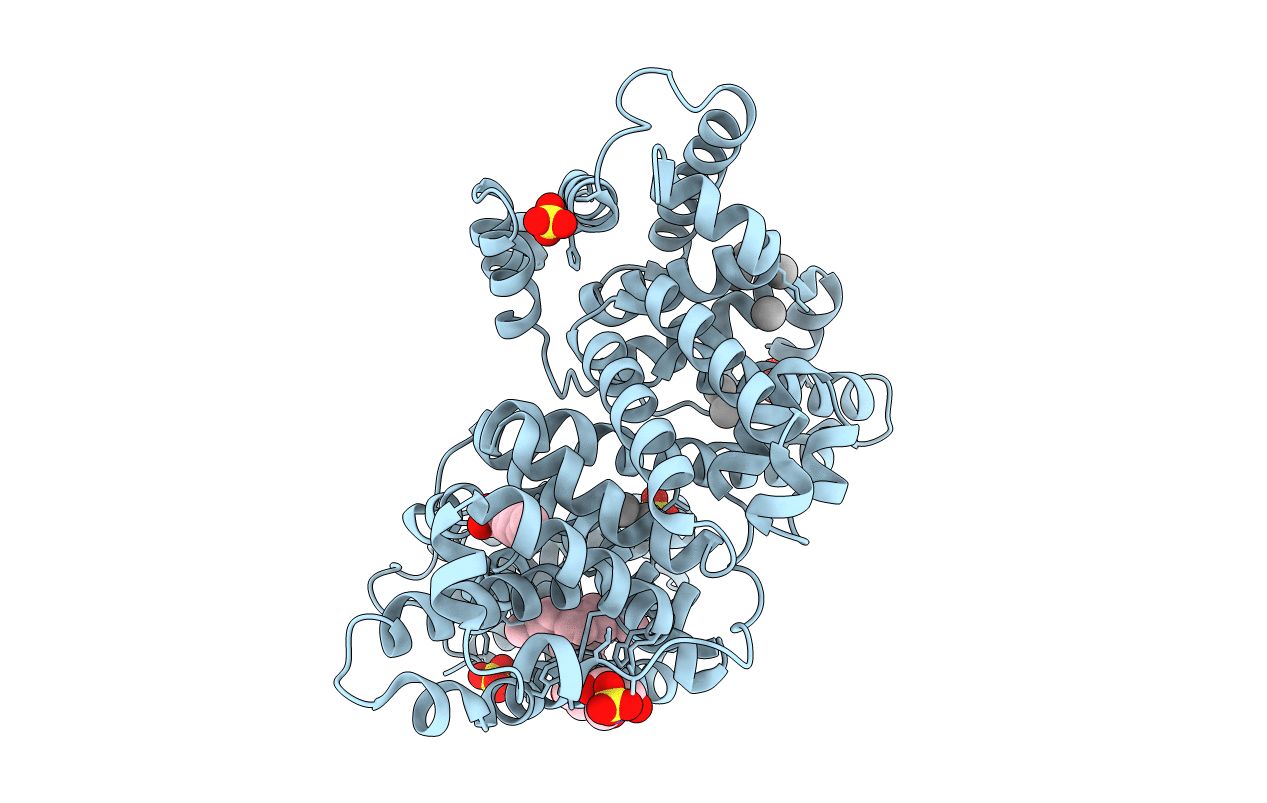
Crystal structure of equine serum albumin in complex with nabumetone
Biological Source:
Source Organism:
Equus caballus (Taxon ID: 9796)
Method Details:
Experimental Method:
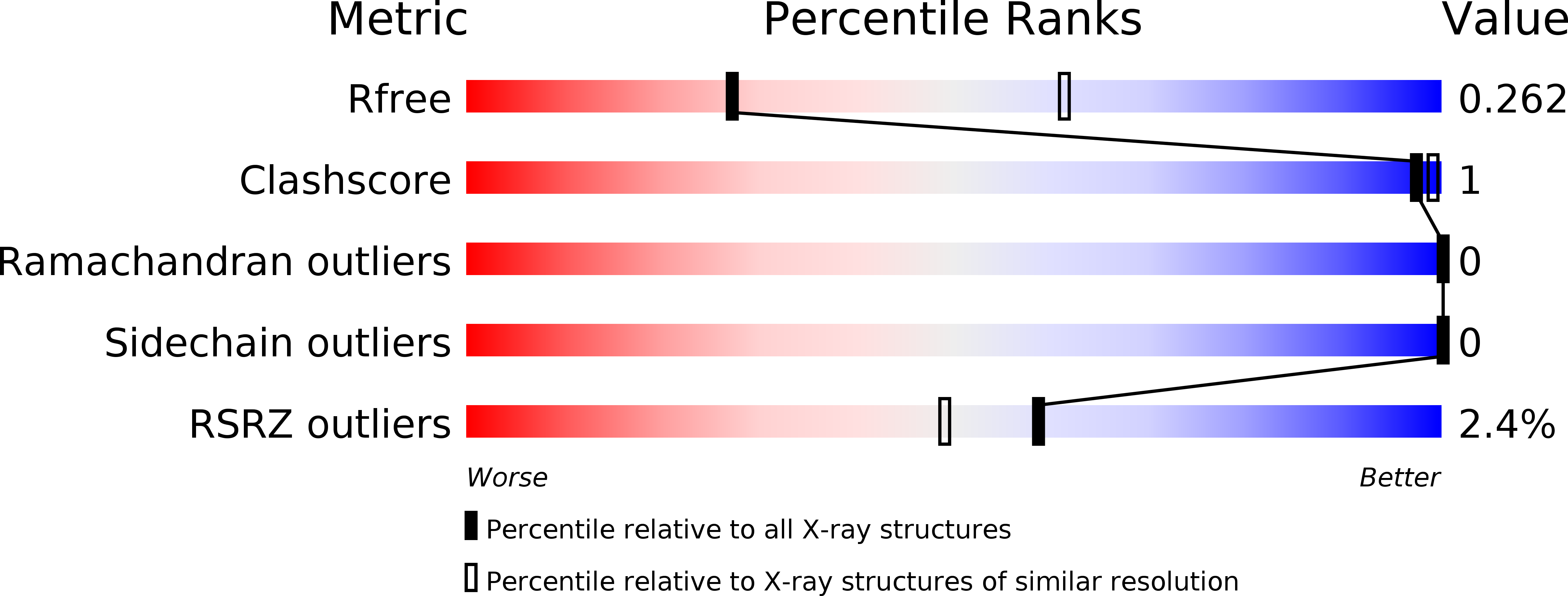
Resolution:
2.80 Å
R-Value Free:
0.25
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 61